Abstract
The Sec34/35 complex was identified as one of the evolutionarily conserved protein complexes that regulates a cis-Golgi step in intracellular vesicular transport. We have identified three new proteins that associate with Sec35p and Sec34p in yeast cytosol. Mutations in these Sec34/35 complex subunits result in defects in basic Golgi functions, including glycosylation of secretory proteins, protein sorting, and retention of Golgi resident proteins. Furthermore, the Sec34/35 complex interacts genetically and physically with the Rab protein Ypt1p, intra-Golgi SNARE molecules, as well as with Golgi vesicle coat complex COPI. We propose that the Sec34/35 protein complex acts as a tether that connects cis-Golgi membranes and COPI-coated, retrogradely targeted intra-Golgi vesicles.
Keywords: Golgi; Rab GTPase; SNARE; coatomer; Sec34/35p complex
Introduction
The sorting and targeting of protein transport vesicles to specific docking and fusion sites is essential for the maintenance of a proper organization of membrane compartments in the eukaryotic cell. To ensure the appropriate directionality of membrane flow, target organelles must possess a molecular machinery allowing for the specific recognition and docking of incoming vesicles. The docking reaction requires a set of integral membrane proteins on the vesicle and target membranes, termed v-SNAREs and t-SNAREs (vesicle- and target membrane–soluble N-ethylmaleimide–sensitive fusion protein attachment protein receptors, respectively). SNARE proteins are thought to confer specificity of vesicle targeting and fusion through their pair-wise interactions (Söllner et al., 1993). However, there is good evidence that SNAREs cannot be the only targeting components: some SNAREs can function at several transport steps in vivo (Fischer von Mollard et al., 1997), SNAREs that faithfully function at only specific transport steps in vivo can interact quite promiscuously in vitro, and some SNAREs have been found in multiple SNARE complexes (Fischer von Mollard et al., 1997; Lupashin et al., 1997; Nichols and Pelham, 1998). In a similar fashion, members of the Rab GTPase family were once thought to be the principle determinants of this targeting specificity based on the fact that distinct family members display unique organellar localizations that correlate with their sites of action (Ferro-Novick and Novick, 1993). However, it has been demonstrated that a single chimeric Rab protein can function at two transport steps (Brennwald and Novick, 1993), indicating also that Rabs cannot be the sole targeting determinants. Given that neither SNAREs nor Rabs, although necessary, seem sufficient to be the sole governors of targeting specificity, other components are likely to play important roles. Additional candidate players include so-called tethering factors, which are proteins that bind membranes together before SNARE interactions occur (Pfeffer, 1999; for review see Waters and Pfeffer, 1999).
A genetic screen for yeast secretory mutants identified temperature-sensitive alleles of two genes, SEC34 and SEC35. Yeast cells harboring these mutations, when incubated at the restrictive temperature, are defective in ER to Golgi complex transport and accumulate large numbers of vesicles (Wuestehube et al., 1996). The sec34 and sec35 mutants can be efficiently suppressed by overexpression of YPT1, which encodes a Rab-like GTPase required early in the secretory pathway. Weaker suppression is also evident upon overexpression of genes encoding the v-SNAREs SEC22, BET1, or YKT6, a trait shared with all previously characterized ER-to-Golgi-complex tethering factors (Sapperstein et al., 1996; VanRheenen et al., 1998, 1999). Based on these data, it was hypothesized that SEC34 and SEC35 might be involved in tethering. Indeed, it could demonstrated that SEC34 and SEC35 genes display a genetic interaction with genes involved in tethering, and that Sec35p is required in this process as revealed by an in vitro assay (VanRheenen et al., 1998, 1999).
The Sec34p has also been described as Grd20p, a protein that when mutated affects the proper localization of yeast enzymes in the TGN (Spelbrink and Nothwehr, 1999). Finally, we have recently shown that Sec34p is evolutionarily conserved by cloning and characterization of a human Sec34p homologue, which is localized to cis-Golgi cisternae and not on the transport vesicles or vesicular–tubular clusters (Suvorova et al., 2001).
It has been shown that the yeast sec34-2 and sec35-1 mutations display a synthetic lethal interaction with each other, a genetic result readily explained by the finding that Sec34p and Sec35p can interact directly in two-hybrid assays. Fractionation of yeast cytosol indicates that Sec34p and Sec35p exist together in a high molecular mass protein complex(es) (Kim et al., 1999; VanRheenen et al., 1999).
To better understand the function of the Sec34/35p complex in vesicle trafficking, we isolated and characterized interacting proteins. In this paper we describe three new subunits of the cytosolic Sec34/35 complex, and demonstrate that mutant cells that are defective in subunits of the Sec34/35 protein complex are compromised in basic Golgi functions, including protein sorting, secretory protein glycosylation, and correct localization of resident Golgi proteins. We also describe the in vivo and in vitro associations of the Sec34/35p complex with a subset of Golgi SNARE proteins and with the retrograde vesicle coat protein complex COPI. The Sec34/35 complex, SNAREs, and COPI may cooperate to provide a targeting system to recruit retrograde intra-Golgi vesicles to the appropriate cis-Golgi cisternae, thus maintaining the proper organization of the Golgi complex.
Results
Identification of three novel subunits of the Sec34/35 protein complex
To identify additional subunits of the Sec34/35 protein complex, we made use of the tandem affinity purification (TAP)* tagging system (Rigaut et al., 1999) that has been used to isolate a number of native protein complexes from yeast (Bouveret et al., 2000; Puig et al., 2001). Homologous recombination was used to insert the TAP tag (two copies of the IgG binding domain from protein A and calmodulin binding domain) at the C terminus of the SEC35 gene product, leading to its stable expression at the endogenous levels in a protease-deficient yeast strain. The Sec35-TAP protein was fully functional, as the TAP-tagged strain had a growth rate indistinguishable from wild-type at all temperatures. Cytosol was prepared from this Sec35-TAP strain, high molecular mass protein complexes were concentrated by ammonium sulfate precipitation, and the resulting concentrate was passed over a human IgG-Sepharose column. The ammonium sulfate step was essential for the Sec34/35 complex purification, as some (∼15%) of Sec35p is present in yeast cytosol as a monomer (unpublished data) and this form competes with the complex upon affinity purification. Bound proteins were cleaved of the IgG-Sepharose by TEV protease and the TEV eluate was incubated with calmodulin-agarose beads. The bound proteins were then eluted by EGTA and analyzed by SDS-PAGE. Fig. 1 A shows that several proteins were isolated from the tagged strain, including a band of the expected size for TAP-tagged Sec35p (Sec35-TAP). The coprecipitating proteins were identified by subjecting the bands to in-gel tryptic digestion and determining the masses of the tryptic fragments by mass spectrometry. This revealed the presence of Sec35-TAP and Sec34p as expected, as well as three previously uncharacterized proteins that had coprecipitated with Sec35-TAP. All of these proteins were absent in identically purified samples prepared from a control strain (unpublished data). The three identified polypeptides are encoded by the yeast ORFs Ygl223c, Ynl041c, and Ypr105c, and their properties were not previously described in the literature. The sequence of Ygl223c predicts a protein of 417 amino acid residues with a molecular mass of 48.3 kD. The systematic disruption of the yeast ORFs performed by the European Network for Functional Analysis (http://www-sequence.stanford.edu/group/yeast_deletion_project/consortium.html) consortium revealed that a ygl223c-null mutation leads to a slow growth phenotype (Lucau-Danila et al., 2000). Ynl041cp is a nonessential ORF (Winzeler et al., 1999) that encodes a hydrophilic 96.9-kD protein, which harbors heptad repeats capable of forming a coiled coil in an N-terminal region (amino acids 158–187, 222–243, and 252–279) as predicted by the COILS algorithm (with a window size 21 and the MTDIK matrix) (Lupas et al., 1991). Finally, Ypr105cp is an essential ORF (Winzeler et al., 1999) encoding a protein of a predicted molecular mass of a 98.6-kD protein with a putative short coiled coil structure in its N-terminal region (amino acids 62–88).
Figure 1.
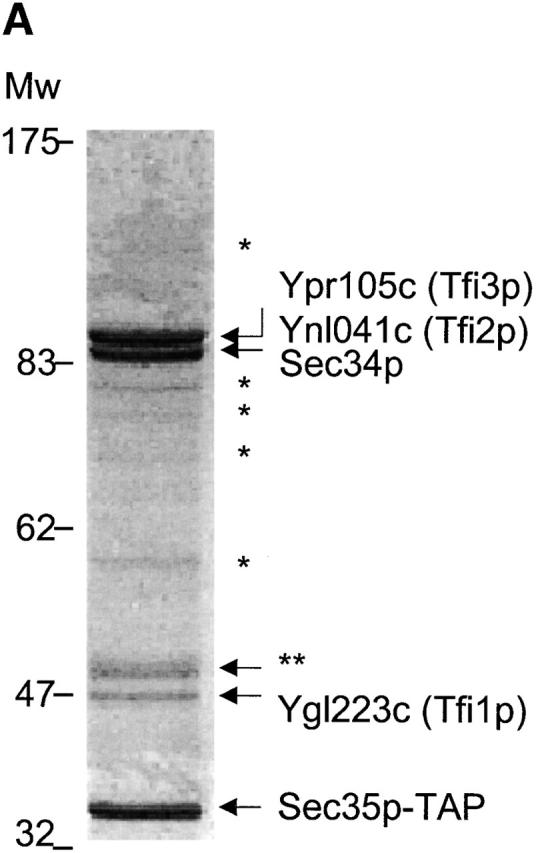
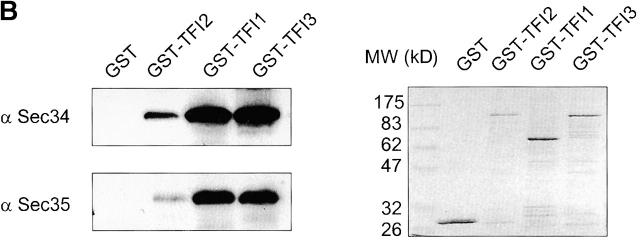
Sec35p-TAP complex purified from yeast cytosol. (A) Sec35p-TAP is associated with four other proteins. Proteins eluted from calmodulin-agarose beads were separated by gel electrophoresis and stained with Coomassie blue. The indicated bands were identified by mass spectroscopy of tryptic fragments. The band labeled with double asterisks represents a proteolytic fragment of Tfi3p. No readable spectra were obtained from the minor bands labeled with asterisks. (B) GST-tagged TFI1, TFI2, and TFI3 interact with both Sec34p and Sec35p. The GST-tagged proteins were expressed in cells of appropriate gene deletion strains, in which the GST-chimera was the only source of TFI1, TFI2, or TFI3 protein. Membrane fractions were obtained after centrifugation at 150,000 g, 1 h, 4°C. Extracted membrane proteins (2 mg) were incubated with 50 μl prewashed glutathione–Sepharose beads. The eluates were loaded on the 10% SDS-PAGE and then analyzed by immunoblot with α Sec34p and α Sec35p (left), or stained with Coomassie blue (right).
To confirm the mass spectrometric identification of the proteins, yeast strains were constructed in which a GST tag was attached to the N terminus of each of the three ORFs, and the resulting plasmids were then transformed into yeast strains that harbored a deletion of the corresponding chromosomal gene. Both Sec34p and Sec35p were found to be present in glutathione–Sepharose precipitations from every strain, with the exception of the control strain (Fig. 1 B). This confirmed that the proteins identified by mass spectrometry are indeed associated with Sec35p, and indicated that all three novel proteins are components of the Sec34/Sec35 protein complex. Because the three novel proteins were identified through their interaction with Sec35-TAP hybrid protein, we named them TFI (thirty-five interacting) proteins: Tfi1p (Ygl223cp), Tfi2p (Ynl041c), Tfi3p (Ypr105c). Interestingly, a recent comprehensive high-throughput two-hybrid analysis of the yeast protein interactome (Uetz et al., 2000; Ito et al., 2001) revealed direct interactions between Sec35p and Tfi2p, Sec35p and Tfi3p, and Tfi1p and Tfi2p. Database searches demonstrated that Tfi2p and Tfi3p have closely related homologues in Caenorhabditis elegans, Drosophila, Arabidopsis, and humans (KIAA1134 for Tfi2p and DKFZP586E1519 for Tfi3p), whereas searches for Tfi1p homologues reveal only one plant (Sorghum bicolor) cDNA sequence (BG463297) that encodes a putatively homologous protein that is 29% identical and 42% similar to Tfi1p.
Tfi3p colocalizes with the Sec34/Sec35 protein complex
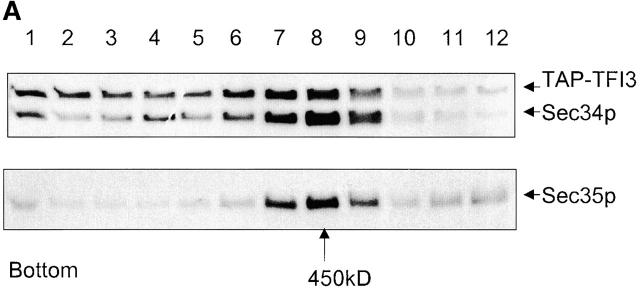
To begin the characterization of TFI proteins, a strain expressing a TAP-tagged version of Tfi3p was generated. Purification and Western blot analysis of Tfi3p-TAP from cytosol prepared from this strain confirmed the physical interaction with Sec34p and Sec35p (unpublished data). To determine what fraction of Tfi3p is associated with Sec34p and Sec35p, cytosol was prepared from the Tfi3p-TAP– tagged strain and loaded on a glycerol velocity gradient. After this separation step, Sec34p, Sec35p, and Tfi3p-TAP were visualized by Western blot (Fig. 2 A), revealing that the major peak of Tfi3p was recovered in fractions 7 and 8 together with Sec34p and Sec35p. Small amounts of Tfi3p and Sec34p were also found in fractions 1–6, and a minor peak of Sec35p was observed in fractions 11 and 12. The latter may represent the Sec35p monomer pool that was previously observed (VanRheenen et al., 1999). No such monomeric Tfi3p was detected in cytosolic fractions. These results are consistent with all Tfi3p existing in a protein complex with Sec34p and Sec35p.
Figure 2.
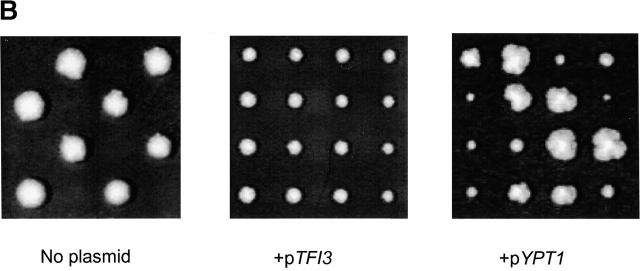
Properties of Tfi3p. (A) Tfi3 protein comigrates with the Sec34/Sec35p complex on a glycerol velocity gradient. Cytosol (0.4 ml) from the LY367 yeast strain was fractionated on a 10–30% (wt/vol) glycerol gradient. Fractions were collected, loaded on 10% SDS-PAGE, and analyzed by immunoblotting using anti-Sec34p and anti-Sec35p antibodies. (B) Deletion of TFI3 leads to lethality, which can be suppressed by expression of YPT1. Left, the tfi1Δ/TFI1 diploid strain was sporulated, tetrads dissected and incubated on YPD plates at 25°C for 6 d. Two representative tetrads are shown, each of which displayed a 2+:2− segregation. (Center) The growth defect of a tfi1Δ strain is complemented by TFI3, as expected. (Right) The growth defect of a tfi1Δ strain is suppressed by overexpression of YPT1. The tfi1Δ/TFI1 diploid strain containing plasmids pTFI3 or pYPT1 was sporulated and subjected to tetrad analysis. Tetrads were incubated on YPD plates for 3 d (center) or 6 d (right).
A recent whole-genome approach to construct yeast strains, each with a precise deletion of a single ORF (Winzeler et al., 1999), revealed that both TFI1 and TFI2 are dispensable for growth, and that deletion of TFI3 is lethal. We have previously shown that strains that carry deletions in either SEC34 or SEC35 display a severe growth defect (VanRheenen et al., 1998, 1999). Here we have reinvestigated the essential nature of TFI3 in our strain background. The heterozygous strain (tfi3Δ::Gm/TFI3) was sporulated and dissected, and the resulting tetrads were incubated on rich media at 28°C. As shown on Fig. 2 B (left), a clear 2 dead:2 alive segregation pattern was observed, even after 6 d of growth, confirming the essential nature of TFI3. The growth defect of the tfi3Δ strain is complemented by a plasmid-bearing TFI3 gene (Fig. 2 B, center). In each tetrad, two segregants were URA+ and were able to grow on the G-418–containing media, indicating the presence of both deletion and the plasmid bearing TFI3, respectively.
One of the common features of membrane tethering systems is their dependence on members of the large family of structurally and functionally related Rab GTPases. Previous analysis of the sec34Δ and sec35Δ strains demonstrated that overexpression of the Rab protein Ypt1p significantly improved the growth of both mutant strains (VanRheenen et al., 1998, 1999). Therefore, we explored whether Ypt1p overexpression could also rescue the growth defect in the tfi3Δ strain. The tfi3Δ/TFI3 diploid strain was transformed with a multicopy YPT1 plasmid, and the resulting diploid was sporulated and subjected to tetrad analysis. Indeed, the presence of pYPT1 was able to rescue the lethal phenotype of the tfi3Δ strain (Fig. 2 B, right). This result implies that the lethality observed for a tfi3Δ strain is directly connected with the Sec34/35 protein complex function. Because a tfi3Δ strain that overexpresses Ypt1p was able to grow, we used this strain in further experiments.
The Sec34/35 complex specifically binds to Ypt1p
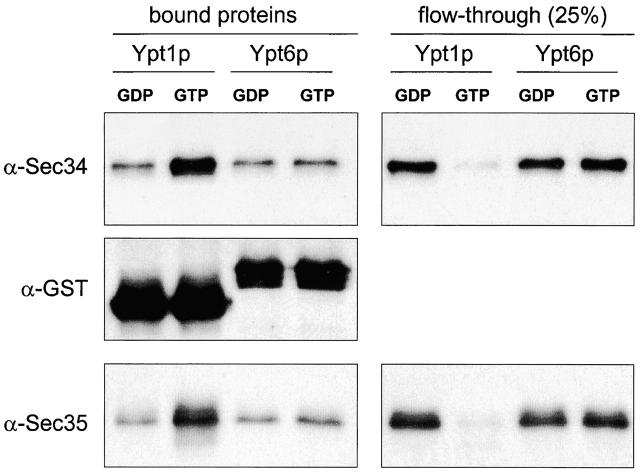
An in vitro binding assay was used to investigate possible physical interactions between Ypt1p and the Sec34/35 complex (Fig. 3). The Sec34/35 protein complex that was purified by the IgG-agarose chromatography from the Tfi3-TAP–bearing yeast strain, was found to be able to specifically bind to GTP-loaded GST-Ypt1p. More than 50% of purified complex bound to the GTP-Ypt1p under the conditions used, as judged by quantitative immunoblot analysis for both Sec34p and Sec35p. The binding of the complex was at least five times less efficient with GDP-loaded GST-Ypt1p, and insignificant with control GST beads (unpublished data). Very low nucleotide-independent binding was obtained with the control GST-Ypt6p (Fig. 3). This correlates with an inability of overexpressed YPT6 to suppress the lethality observed for tfi3Δ and sec34Δ strains (unpublished data). Taken together, i.e. the novel physical interaction of the Sec34/35p complex with GTP-Ypt1p as well as the strong genetic interactions between YPT1 and genes of the Sec34/35 complex subunits (Fig 4; VanRheenen et al., 1998, 1999), we propose that the Sec34/35 complex, like other vesicle tethering factors (Christoforidis et al., 1999; Allan et al., 2000) may operate as a Rab (Ypt1) effector.
Figure 3.
The Sec34/35 complex is an effector of the Ypt1 protein. GST-Ypt1 or GST-Ypt6p (1 μg each), were preloaded with GDP or GTP as indicated, and bound to glutathione–Sepharose beads. Beads were incubated with 0.2 μg of purified Sec34/35 complex in 0.2 ml of binding buffer (40 mM Hepes pH 7.0, 150 mM KoAc, 2 mM MgOAc, 5% glycerol, 1 mM DTT) for 3 h at 4°C. Beads were washed with binding buffer and proteins were eluted with the sample buffer. Eluted proteins and 25% of the unbound material were loaded on 10% SDS-PAGE and immunoblotted with affinity-purified antibodies to GST, Sec34p, and Sec35p.
Figure 4.
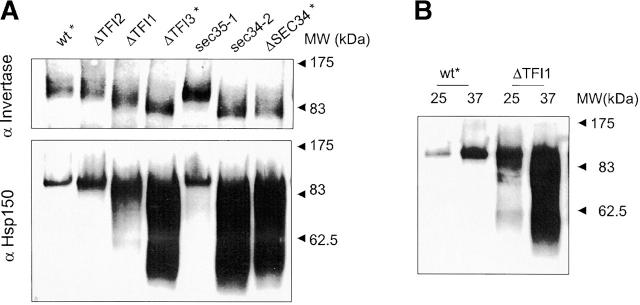
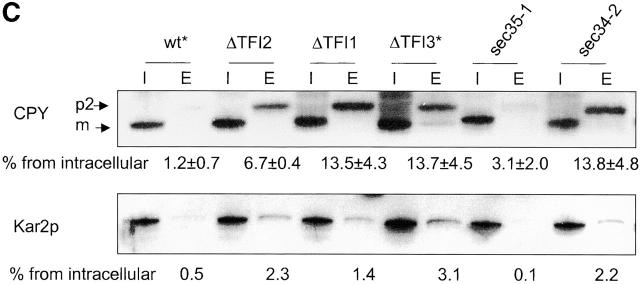
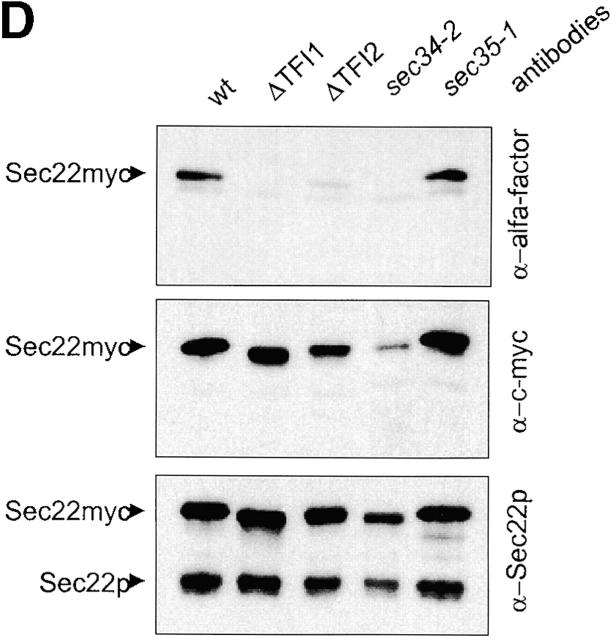
Sec34/35 protein complex mutants are defective in Golgi glycosylation (A and B), CPY sorting (C), and recycling of resident cis-Golgi proteins (D). (A and B) Analysis of glycosylated proteins secreted by different mutants. (A) Wild-type and mutant cells were grown in YPD medium at 25°C for 18 h. The culture medium was collected and proteins concentrated by TCA precipitation. Equivalents of 100 μl of medium were loaded on 10% SDS-PAGE and Western blotted with either anti-invertase (top) or anti-Hsp150 (bottom) antibodies. (B) To analyze HSP150 secretion at the nonpermissive temperature, cells grown overnight at 25°C were washed once with water, resuspended in fresh prewarmed YPD, and incubated for 3 h at 37°C. Proteins from the culture medium were concentrated by TCA precipitation and analyzed as in A. (C) Intracellular (I) and extracellular (E) fractions were prepared from cells grown in YPD medium at 25°C for 18 h. An equivalent of 10% cell lysate and 100% of culture medium were loaded on 10% SDS-PAGE and immunoblotted with either anti-CPY or anti-Kar2 antibodies. Secretion of these proteins was quantified using the Scion Image program and the average of three independent experiments was calculated. Asterisks indicate strains that had been transformed with pYpt1. (D) v-SNAREs are not maintained in the cis-Golgi compartment in Sec34/35 complex mutants. Sec22-myc–α hybrid protein is processed by the late Golgi protease Kex2p in Sec34/35 protein complex defective strains. Immunoblot analysis of v-SNARE–myc–α proteins produced in Sec34/Sec35 protein complex mutants and RSY255 wild-type cells transformed with pWB-Acycα is shown. Aliquots of cells after an overnight incubation at 25°C in selective medium were harvested, and proteins were analyzed by immunoblotting with different antibodies as indicated. A polyclonal anti-Sec22 serum was used to detect the v-SNARE–derived hybrid proteins (uncleaved and cleaved by Kex2p protease) and the endogenous Sec22p. Monoclonal anti–c-myc antibodies were used to detect the Sec22-derived hybrid protein. Anti–α-factor antibody was used to detect only uncleaved hybrid protein.
Sec34/35 complex mutants are defective in Golgi functions
It has been reported previously that sec34-2 and sec35-1 mutant cells accumulate the p1 form of the vacuolar enzyme carboxypeptidase Y (CPY) at restrictive temperature (Wuestehube et al., 1996; VanRheenen et al., 1998, 1999). At the same time, it was noted that some sec34 mutants and sec34Δ cells secrete both the p2 form of CPY and underglycosylated invertase when incubated at temperatures that are semipermissive for growth (Spelbrink and Nothwehr, 1999). To understand the primary trafficking defects that could be observed in Sec34/35 complex mutant cells, we investigated protein glycosylation and CPY sorting at 25°C, a permissive temperature for growth of all mutants used (Fig. 4). At this temperature, all mutants secreted invertase and HSP150 into the culture medium at levels equal to or greater than wild-type cells (Fig. 4 A). Secreted invertase is a heavily N-glycosylated enzyme (Huffaker and Robbins, 1983), and HSP150 is a heavily O-glycosylated protein that is secreted into the culture medium (Lupashin et al., 1992; Russo et al., 1992). The gel mobility of the invertase and HSP150 was dramatically affected in tfi1Δ, tfi3Δ, sec34Δ, and sec34-2 cells, indicating that both N- and O-linked Golgi glycosylation is greatly reduced. The gel mobility of secretory proteins was only slightly affected in tfi2Δ and sec35-1 cells at 25°C.
We have investigated putative HSP150 glycosylation defect in tfi1Δ strain further (Fig. 4 B). The gel mobility of HSP150 secreted by tfi1Δ strain at 25°C was only slightly altered as compared with wild-type, whereas at 37°C, the mobility of HSP150 was significantly increased, a feature that probably reflects a more severe glycosylation defect in the tfi1Δ strain at the elevated temperature. HSP150 expression was stimulated at the high temperature in both wild-type and tfi1Δ cells, but in wild-type cells the gel mobility of the protein was not effected. No intracellular accumulation of HSP150 was observed in either of the strains at both 25 and 37°C (unpublished data).
All Sec34/35 complex mutants are defective in the intracellular sorting of CPY and secrete the p2 form of CPY into the culture medium (Fig. 4 C, top lane). CPY secretion was especially significant in tfi1Δ, tfi3Δ, sec34Δ, and sec34-2 cells (∼13% of total enzyme). Small amounts of mature CPY were also detected in the culture medium from tfi3Δ and sec34Δ cells. This was most probably a result of cell lysis, as a comparable leakage of the ER marker Kar2p (Fig. 4 C, bottom lane) and the cytoplasmic marker GDI (unpublished data) into the culture medium was also detected in tfi3Δ and sec34Δ cells. As expected, wild-type cells used as controls correctly localized CPY to the vacuole (Fig. 4 C).
We also examined the retention/recycling of cis-Golgi v-SNARE proteins, namely Sec22p and Bos1p. Both v-SNAREs act in the fusion of ER-derived vesicles with cis-Golgi membranes, and normally do not travel to the trans-Golgi compartment (Ballensiefen et al., 1998; Ossipov et al., 1999). As described previously (Ballensiefen et al., 1998), α-factor fused to these v-SNAREs through a linker containing a Kex2 cleavage site is a suitable tool for analyzing the targeting of Sec22p and Bos1p. Several Golgi-to-ER recycling mutants exhibit mislocalization of cis-Golgi v-SNAREs, as measured by the cleavage of this reporter by the late Golgi protease Kex2p (Ballensiefen et al., 1998; Ossipov et al., 1999; Andag et al., 2001). Fig. 4 D shows the steady-state level of processing of the Sec22-α−factor chimera in wild-type and Sec34/35 mutant strains incubated at 25°C, as assessed by immunoblotting. It is evident that >90% of Sec22-α proteins was cleaved by Kex2 in tfi1Δ, tfi2Δ, and sec34-2 cells, whereas very little was cleaved by Kex2p in wild-type and sec35-1 mutant cells. Similar results were observed for the Bos1-α reporter construct (unpublished data). Based on these experiments, we conclude that several Sec34/35 complex mutants are defective in the recycling of cis-Golgi v-SNAREs.
The Sec34/35 complex directly interacts with intra-Golgi SNAREs
Previous genetic analysis of the sec34 and sec35 mutations showed that they display genetic interactions with genes encoding SNARE proteins that participate in the docking and fusion of ER-derived vesicles with cis-Golgi membranes (VanRheenen et al., 1998, 1999). To explore potential physical interactions, we tested whether the intact Sec34/35 complex or its individual subunits are capable of interacting directly with the cis-Golgi SNARE machinery.
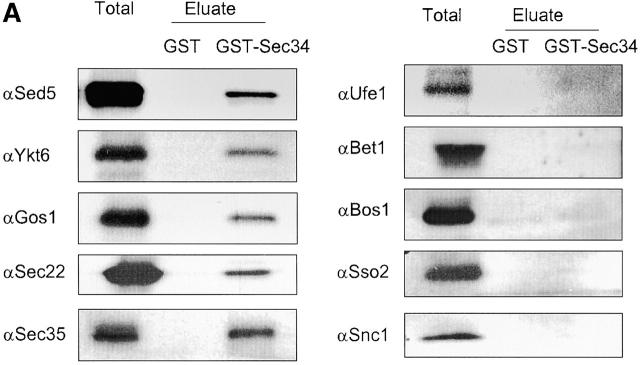
To identify yeast SNARE proteins that may interact with the Sec34/35 complex in vivo, we constructed a yeast strain that expresses GST-Sec34p as the sole source of Sec34 protein. Glutathione–Sepharose affinity chromatography was employed to purify GST-Sec34p and any associated proteins from a solubilized membrane fraction. As shown in Fig. 5 A, Sec35p was recovered together with GST-Sec34p, indicating that our purification conditions were suitable for the efficient recovery of the intact Sec34/35 complex. We find that Sec34p interacts with the cis-Golgi t-SNARE Sed5p and several v-SNAREs that are involved in trafficking to or from the cis-Golgi: Gos1p, Ykt6p, Sec22p (Fig. 5 A, left column), and Vti1p (unpublished data). However, we did not observe stable interactions with two other v-SNAREs, which also act on the same trafficking step, namely Bet1p and Bos1p. Further, no interactions were detected with the ER t-SNARE Ufe1p or the plasma membrane SNAREs Snc1p and Sso2p (Fig. 5 A, right column), or the endosomal t-SNARE Pep12p (unpublished data), confirming the specificity of these interactions. About 10% Sec34p, 3% Sec35p, and 0.1% of Golgi SNARE proteins were specifically recovered on glutathione–Sepharose beads. The low but specific recovery of SNARE proteins may indicate the transient nature of interactions between the Sec34/35 protein complex and the SNARE fusion machinery.
Figure 5.
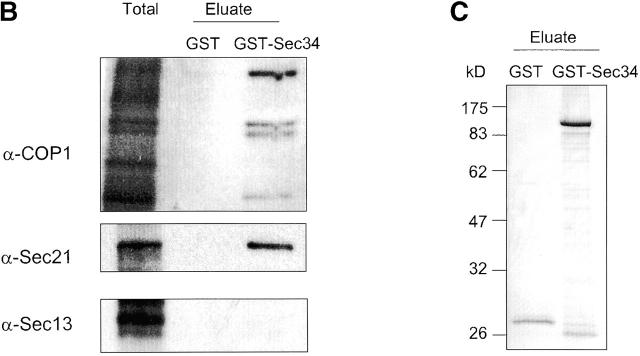
Sec34p interacts with retrograde Golgi SNAREs and with COPI. GST-Sec34p was expressed in a Δsec34 yeast strain. Affinity chromatography on glutathione–Sepharose beads was employed to purify GST-Sec34p and associated proteins from a P100 fraction (Total) that was solubilized in CHN buffer (20 mM Hepes, pH 7.4, 1% CHAPS, 0.15 M NaCl). The beads were eluted with 10 mM glutathione (Eluate). As a control, the GST protein was expressed and purified from the membrane fraction of sec34/Δsec34 strain. The GST-Sec34p– and Sec34-associated proteins were identified by immunoblotting with antibodies to A, SNARE proteins and Sec35p; (B) anti-COPI, Sec21p, or Sec13p. (C) Coomassie blue staining of the GST and GST-Sec34p eluates.
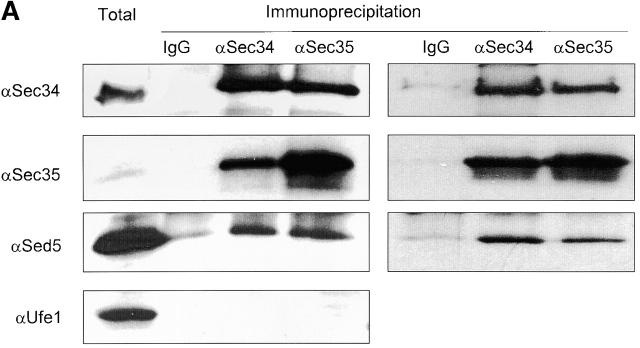
To confirm the interaction of the Sec34/35 complex with SNAREs in GST-independent experiments, we performed native immunoprecipitation of protein complexes from solubilized membrane fractions using affinity-purified antibodies to different components of the cis-Golgi vesicle docking machinery. Indeed, antibodies to either Sec34p or Sec35p coimmunoprecipitated Sed5p (Fig. 6 A), and, in the reciprocal experiment, Sec34p was detected in the Sed5p immunoprecipitate (unpublished data).
Figure 6.
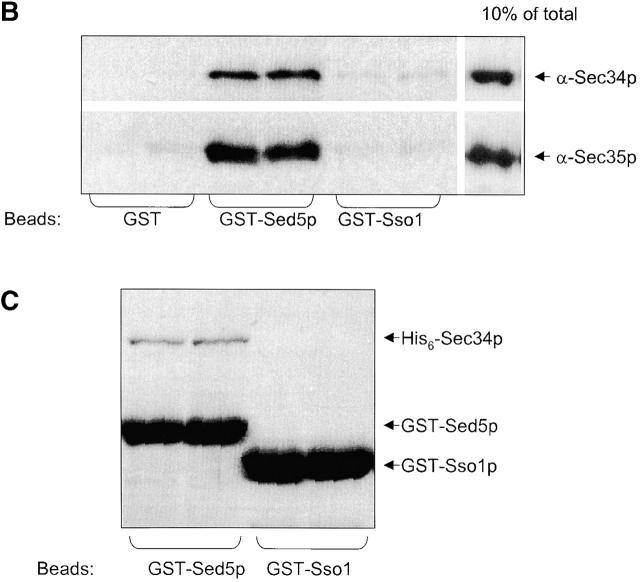
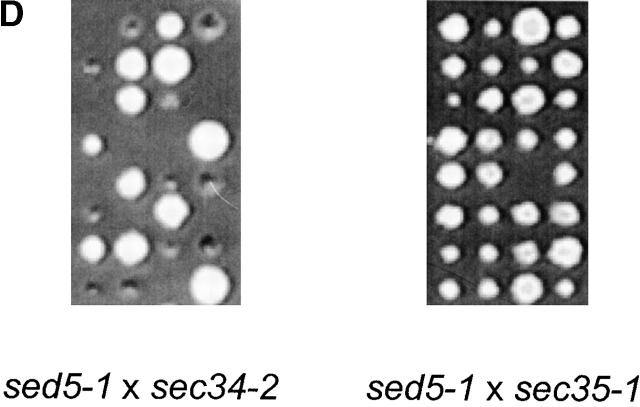
Sec34p and Sed5p interact genetically and physically. (A) Coimmunoprecipitations of putative partners of the Sec34/35 complex. A P100 membrane fraction (Total) from RSY1157 was solubilized in 20 mM Hepes-KOH, pH 7.4, with 0.1 M NaCl and 1% CHAPS. The extract was incubated with protein A Sepharose beads, to which different primary antibodies had been crosslinked. Rabbit IgGs were used as a control for unspecific binding. After washing of the beads, specifically bound proteins were eluted, run on 11%SDS-PAGE, and analyzed by immunoblotting. Two representative experiments performed under identical conditions are shown. Membranes (3% of total) were loaded on the first lane. Approximately 15% of Sec34p and 40% of Sec35p were recovered in the corresponding IP. Approximately 0.2% of Sed5p was specifically coprecipitated in both Sec34 and Sec35 IP's. (B) The Sec34/Sec35 complex interacts with Sed5p in vitro. Purified GST, GST-Sed5p, or GST-Sso1p (5 μg each) were mixed with 0.2 μg of purified Sec34/35 complex in 0.5 ml binding buffer, incubated for 3 h at 4°C, and centrifuged at 20,000 g for 10 min. The supernatant (0.45 ml) was incubated for 1 h at 4°C with 20 μl glutathione–Sepharose beads in the same buffer. Beads were washed and bound proteins were eluted with 10 mM glutathione, separated by 10% SDS-PAGE, and immunoblotted with affinity-purified antibodies to Sec34p and Sec35p. (C) Physical in vitro interaction of Sec34p with Sed5p. Purified proteins (as in B) were bound to glutathione–Sepharose beads, mixed with 5 μg of His6-Sec34p in 0.5 ml of binding buffer and incubated for 3h at 4°C with rotation. Beads were washed and bound proteins were eluted with 10 mM glutathione, separated by 11% SDS-PAGE and stained with Coomassie blue. Approximately 10% of His6-Sec34p were recovered with GST-Sed5p bound to glutathione–Sepharose beads under conditions used. (D) The sed5–1 and sec34–2 alleles display a synthetic lethal interaction. Diploid strains resulting from the mating of GWY234 (sed5–1) with GWY95 (sec34–2) (top) or GWY235 (sed5–1) with GWY93 (sec35–1) (bottom) were sporulated, tetrad dissected, and incubated on YPD plates at 30°C for 3 d. Eight representative tetrads for each dissection are shown.
To determine whether the in vivo interaction of the Sec34/Sec35 protein complex with Sed5p was direct, we attempted to reconstitute this interaction with purified proteins (Fig. 6, B and C). Purified GST, GST-Sed5p, or GST-Sso1p was mixed with purified Sec34/35 complex, and complexes formed were recovered on glutathione–Sepharose beads. An efficient interaction between Sed5p and Sec34/Sec35 protein complex was observed (Fig. 6 B). The interaction was specific, as no Sec34p or Sec35p was recovered with either GST or GSP-Sso1p loaded beads.
Data from the immunoprecipitation experiments (Fig. 6 A) indicated that the amount of coprecipitated Sed5p was proportional to the amount of isolated Sec34p, suggesting that Sed5p and Sec34p may interact directly. To test this hypothesis, His6-tagged Sec34p (His6-Sec34p) was incubated with GST-Sed5p or GST-Sec35p, and complexes formed were recovered on glutathione–Sepharose beads. GST-Sso1p served as a negative control. His6-Sec34p did not bind to GST-Sso1p (Fig. 6 C, right lanes). A physical interaction between Sec34p and Sec35p was demonstrated previously using the yeast two-hybrid assay (VanRheenen et al., 1999), and indeed His6-Sec34p could bind to GST-Sec35p in a concentration-dependent manner (unpublished data). Most importantly, His6-Sec34p also bound efficiently (∼10% of the input) to GST-Sed5p (Fig. 6 C, left lanes). This result corroborates the Sec34/35 complex-Sed5p interaction observed in vivo and in vitro and demonstrates that the Sec34p-Sed5p interaction is direct.
The genetic interactions of SEC34 and SEC35 with genes encoding v-SNARE proteins involved in the docking stage of vesicular transport reported earlier (VanRheenen et al., 1998, 1999) further lead us to examine whether mutations in the two genes would display synthetic lethal interactions with a mutation in SED5. To do this, we generated two diploid strains heterozygous for either the sec34-2 and sed5-1 alleles or the sec35-1 and sed5-1 alleles and subjected them to tetrad analysis at 25 and 30°C. Although these temperatures are permissive for growth for all three mutant haploid strains, tetrads from the diploid sed5-1/SED5 SEC34/sec34-2 strain yielded numerous inviable colonies at either temperature (Fig. 6 D, left panel). Examination of the viable segregants in each tetrad for temperature sensitivity revealed a pattern in which the inviable segregants are predicted to be the double mutants, combining both the sec34-2 and the sed5-1 alleles. In contrast, tetrads from the diploid sed5-1/SED5 SEC35/sec35-1 strain yielded viable spores (Fig. 6 D, right panel). It is important to note that sec35-1 strain is more temperature sensitive as compared with the sec34-2 strain (unpublished data). Therefore, the sed5-1 mutation displays a gene-specific synthetic lethal interaction with the sec34-2 mutation, but not with the sec35-1 mutation.
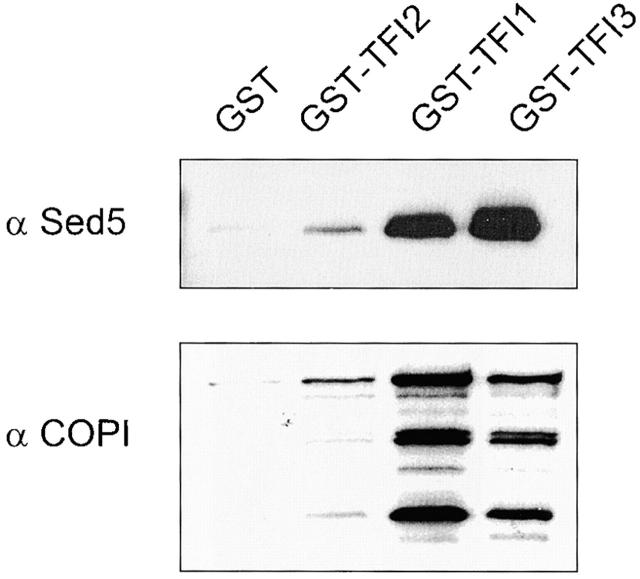
To test the idea that the whole Sec34/35 protein complex may interact with SNARE molecules, we used yeast strains that expressed a GST-tagged version of Tfi1p, Tfi2p, or Tfi3p. Indeed, Sed5p (Fig. 7, top lane) was found to be present in glutathione–Sepharose precipitations from every strain, with the exception of the control strain.
Figure 7.
GST-tagged TFI1, TFI2, and TFI3 interact with both Sed5p and COPI proteins. GST-tagged proteins were expressed in the cells of appropriate gene deletion strains, in which the GST-chimera was the only source of TFI1, TFI2, or TFI3 proteins. A membrane fraction was obtained after centrifugation at 150,000 g for 1 h at 4°C. Extracted membrane proteins (2 mg) were incubated with 50 μl prewashed glutathione–Sepharose beads. Each eluate was loaded on the 10% SDS-PAGE and analyzed by immunoblot with α-Sed5p and α-coatomer sera. The Coomassie blue staining of the samples presents on the left panel of the Fig. 1 B.
The Sec34/35 complex interacts with COPI
To obtain additional clues as to the involvement of the Sec34/35 complex in cis-Golgi retrograde and/or anterograde vesicle trafficking, we investigated possible interactions of Sec34/35 protein complex components with vesicle coat proteins involved in these trafficking steps. First, we employed glutathione–Sepharose affinity chromatography to purify GST-Sec34p and associated proteins from solubilized membrane fractions from yeast cells. After washing the beads to remove nonspecifically bound proteins, antibodies were used to monitor the binding of two different vesicle coats to GST-Sec34p. Anti-coatomer and anti-Sec21p (γ-COPI) resulted in a strong signal (Fig. 5 B, right lane), whereas no signal was obtained with antibodies to the COPII component Sec13p. The signals were specific for the Sec34 part of the fusion protein, as no binding was observed when lysates from cells expressing GST alone were analyzed (Fig. 5 B, middle lane). Not only GST-Sec34, but the whole Sec34/35 complex interacts with coatomer, as we were able to detect COPI in GST-Tfi1p, GST-Tfi2p, and GST-Tfi3p pulldowns from extracts of detergent-lysed yeast cells (Fig. 7, bottom).
To verify and extend these findings, we incubated extracts of detergent-lysed yeast cells with GST-Sec35p fusion protein purified from Escherichia coli. In line with the results obtained with GST-Sec34p expressed in yeast, COPI showed a strong specific binding to GST-Sec35p (unpublished data).
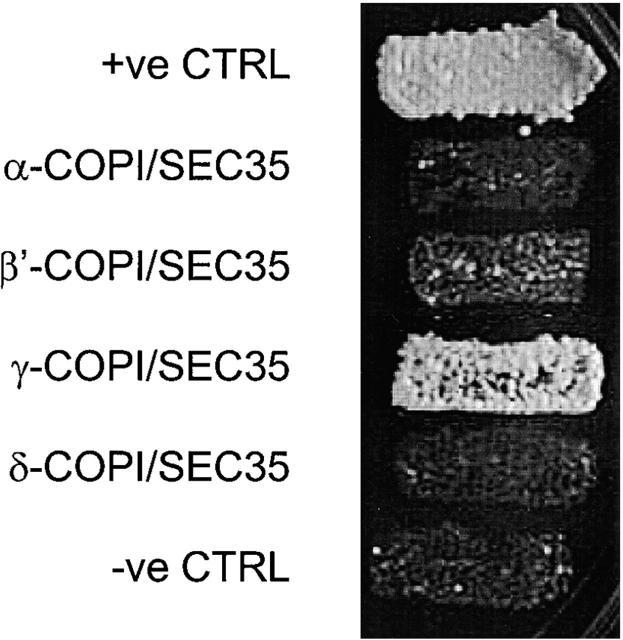
In order to understand the Sec34/35 protein complex interaction in more detail, we used the two-hybrid system to test for possible direct interactions of coatomer subunits with Sec34p and Sec35p. Using the sensitive LexA two-hybrid system (Golemis and Khazak, 1997), we find that yeast γ-COP (Sec21p) interacts specifically with Sec35p (Fig 8). We have also tried the three new ORFs both as bait or as prey against the COPI subunits in the LexA two-hybrid system, but unfortunately, as bait, all three ORFs displayed intrinsic activation, i.e. unusable background (unpublished data).
Figure 8.
SEC35 interacts directly with γ-COP in the two-hybrid system. Full-length SEC35 in the prey vector was tested against coatomer subunits in the bait vector. An interaction between β-COP (bait) and δ-COP (prey) served as an internal positive control. Results shown are from the growth assay on minimal plates lacking leucine in reporter strain EGY48 (see Materials and methods for details). Note that SEC35 interacts with γ-COP. No interactions with coatomer subunits were detected for SEC34 (unpublished data).
Next, we examined whether mutations in the SEC34 or SEC35 genes might display synthetic lethal interactions with a mutation in SEC21. To do this, we generated two diploid strains heterozygous for either both the sec34-2 and sec21-1 alleles or the sec35-1 and sec21-1 alleles, and subjected them to tetrad analysis. Although all three mutant haploid strains are permissive for growth at 25, 30, and 33°C, tetrads from both diploid strains yielded numerous colonies that were unable to grow at 30°C (unpublished data). Examination of the segregants in each tetrad for temperature sensitivity revealed a pattern in which the segregants inviable at 30°C are predicted to be the double mutants. We conclude that sec21-1 displays not a very strong, but a specific synthetic lethal interaction with both the sec34-2 and sec35-1 mutations.
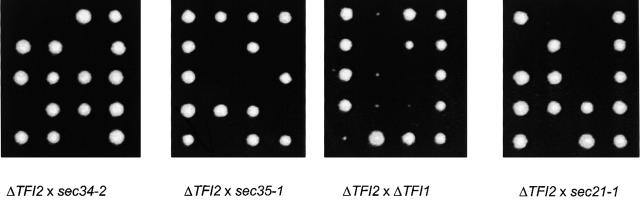
In accordance with the results above, tetrad analysis after sporulation of the diploid tfi2Δ/TFI2 SEC21/sec21-1 strain also yielded numerous inviable colonies (Fig. 9, right). Again, examination of the viable segregants in each tetrad for temperature sensitivity revealed a pattern in which the inviable segregants are predicted to be those containing both the tfi2Δ and the sec21-1 alleles. Thus, although the parent haploid strain that carries the tfi2Δ allele is not temperature sensitive for growth per se, it displays strong genetic interactions with mutations in genes encoding other subunits of the Sec34/35 protein complex, as well as with a mutation in the γ-COP encoding gene, SEC21 (Fig. 9). This result corroborates the Sec34/35 complex–COPI interaction observed in vivo.
Figure 9.
The TFI2 gene product is essential in cells that also harbor mutations in other subunits of the Sec34/35 complex or in the SEC21 gene encoding γ-COP. Diploid strains resulting from the mating of LY240 (tfi2Δ) with either GWY95 (sec34–2), GWY93 (sec35–1), LY241 (tfi1Δ), or RSY277 (sec21–1) were sporulated, tetrads dissected, and incubated on YPD plates at 30°C for 3 d. Five representative tetrads for each dissection are shown.
Discussion
In this study, we used tandem affinity chromatography and subsequent mass spectroscopy analysis of purified proteins to identify three new proteins, Tfi1p, Tfi2p, and Tfi3p, that are associated with Sec35p in yeast cytosol. We have shown that these proteins physically and functionally interact with both Sec35p and Sec34p, and thus, most likely, are subunits of the Sec34/35 vesicle tethering complex. Furthermore, we characterize their interactions with SNAREs, the small GTPase Ypt1p and COP I vesicle coat proteins.
While this paper was in preparation, Whyte and Munro (2001b) published a report on the purification and characterization of six new proteins that interact with Sec34 and Sec35. These proteins were found through their association with the novel Dor1 protein, which was isolated from detergent-solubilized yeast extracts. Three of the Dor1p-associated proteins are identical to the Tfi proteins identified here, namely Tfi1p = Cod3p, Tfi2p = Cod2p, and Tfi3p = Cod1p. In addition to these three common proteins, Whyte and Munro (2001b) found three additional polypeptides, Dor1p, Cod4p, and Cod5p, that were not detectable in the Sec35p-TAP complex isolated in our experiments (unpublished data). It is possible that through our isolation procedure we have purified only the stable soluble core of Sec34/35 protein complex that exists in yeast cytosol. The large eight-subunit complex may be formed on the surface of the Golgi membrane, in which case it could be purified only from detergent-lysed membranes. In accordance with such a model, previous gel filtration analysis of the Sec34/35 complex indicated two different sizes: ∼480 kD (Kim et al., 1999) or up to 750 kD (VanRheenen et al., 1999). However, it also remains possible that there are in fact two or more classes of Sec34/35 complex present in the yeast cell.
Examination of the properties of one of the novel proteins, Tfi3p, revealed that upon fractionation of yeast cytosol on glycerol velocity gradients, Tfi3p comigrated with both Sec34p and Sec35p. No pool of monomeric soluble Tfi3p was detected. In addition, Tfi3p, like Sec34p and Sec35p, exists in yeast cell in both soluble and membrane-bound pools (unpublished data). TFI3 is an essential gene, but the lethality in a TFI3 deletion strain can be partially rescued by high-level expression of YPT1, an interesting phenotype that the tfi3Δ strain shares with the ΔSEC34 and ΔSEC35 strains in which YPT1 overexpression also rescue the gene deletion (VanRheenen et al., 1998, 1999).
The Sec34/35 complex interacts with Ypt1p not only genetically but also physically. Like other vesicle tethering factors and complexes described previously (Christoforidis et al., 1999; Allan et al., 2000; Seals et al., 2000), the Sec34/35p complex may thus operate as an effector of this Rab-type small GTPase. We have demonstrated here that the Sec34/35p complex specifically binds to the activated form of Ypt1p in an in vitro binding assay. Purified Sec34p or Sec35p alone were unable to bind Ypt1p in vitro (unpublished data), perhaps suggesting that one of the novel Sec34/35 complex subunits, rather than Sec34p or Sec35p themselves, serves as the link between the complex and membrane-associated Rab protein. Further experiments will address this question.
To further define the role of the Sec34/35 complex in protein trafficking, we investigated possible primary protein trafficking defects associated with the loss of Sec34/35 complex function. Three of the complex subunits, Sec34p, Sec35p, and Tfi3p are essential for normal cell growth, as deletion of the genes encoding them leads to a phenotype of extremely slow growth, even at the most permissive temperature (VanRheenen et al., 1998, 1999; Winzeler et al., 1999). In a contrast, a deletion of TFI1 only leads to minor defects in growth at 25°C; a Δtfi2 strain grows like wild-type at 25 or 30°C and shows only very subtle growth defects at elevated temperatures (unpublished data). Analysis of trafficking defects in these mutants when grown at permissive temperature revealed no defects in secretion—both markers tested, invertase and HSP150, were secreted from cells at levels comparable to wild-type cells. Surprisingly, all mutant strains tested were defective for the efficient delivery of CPY to the vacuole, which instead mislocalized this vacuolar enzyme to the culture medium. In addition, cells mutated for TFI1, TFI3, or SEC34 were defective in both N- and O-glycosylation reactions associated with the Golgi apparatus. At restrictive temperatures, both sec34-2 and sec35-1 mutant cells have been shown to accumulate the p1 precursor form of CPY lacking Golgi-specific sugar modifications (Wuestehube et al., 1996). The aberrant glycosylation of secretory proteins observed here suggests that there may be a defect in recycling of vesicles that carry outer-chain glycosyltransferases back to their proper compartment within the Golgi. In line with this observation, overexpression of the cis-Golgi mannosyltransferase Och1p was able to partially rescue the slow growth phenotype in sec34-2 and sec35-1 mutant strains (unpublished data). Furthermore, and also consistent with a general defect in intra-Golgi recycling, the correct localization (achieved by recycling and/or retention) of the cis-Golgi resident v-SNARE proteins Sec22p and Bos1p is affected in cells that carry mutations or deletions in Sec34/35 complex components.
Importantly, Golgi-localized Ypt1p was previously shown to operate on several vesicle trafficking steps. This Rab protein is required not only for the fusion of ER-derived anterograde vesicles with cis-Golgi membranes (Rexach and Schekman, 1991; Lupashin et al., 1996), but also for the subsequent step(s) in intra-Golgi trafficking (Jedd et al., 1995). Moreover, genetic interactions observed between Arf GEFs and Ypt GTPases suggest the existence of a Ypt-Arf GTPase regulatory cascade in the secretory pathway (Jones et al., 1999). Furthermore, overexpression of YPT1 suppresses the growth defects of a temperature-sensitive yeast strain that expresses defective Ypt6 protein (Li and Warner, 1998), another yeast Rab family member involved in the retrograde intra-Golgi trafficking (Bensen et al., 2001), thus implicating Ypt1p itself in the regulation of the retrograde trafficking pathway.
A direct involvement of the Sec34/35 protein complex in intra-Golgi retrograde trafficking is supported by our protein–protein interaction data. We have shown here that the Sec34/35 protein complex in vivo interacts with a subset of yeast SNARE molecules which includes Sed5p, Sec22p, Ykt6p, Gos1p, and Vti1p. All of these SNAREs are implicated in the retrograde trafficking pathway (Banfield et al., 1995; Lupashin et al., 1997; von Mollard et al., 1997; Bensen et al., 2001). We have also demonstrated that Sec34p and Sed5p can efficiently interact with each other in vitro, and that mutations in the two genes display a synthetic lethal interaction. Other vesicle tethering factors have been reported to interact with SNARE proteins on target membranes: the mammalian exocyst was shown to coprecipitate with the plasma membrane t-SNARE syntaxin (Hsu et al., 1996), and EEA1, an endosomal tether that display significant sequence similarity to hSec34p (Suvorova et al., 2001), interacts directly with syntaxin-6 (Simonsen et al., 1999).
Although the temperature-sensitive phenotype of the sec34-2 and sec35-1 mutants can be partially suppressed by overexpression of the ER-Golgi v-SNARE Bet1p (VanRheenen et al., 1998; VanRheenen et al., 1999), we failed to detect any protein–protein interaction between this anterograde SNARE molecule and the Sec34/35 complex. The same negative result was obtained with another ER-Golgi anterograde SNARE, Bos1p. It is important to note that the in vitro defects of sec34-2 and sec35-1 mutants in the tethering of anterograde ER-derived vesicles were observed only at a restrictive (29°C) temperature (VanRheenen et al., 1998, 1999), whereas the Golgi-associated protein glycosylation and sorting defects are strikingly evident even at permissive temperature. Taken together, these results suggest that the Sec34/35p complex is primarily involved in the retrograde intra-Golgi trafficking pathway, and that the effects of mutations in SEC34 and SEC35 on anterograde ER to Golgi transport may only be indirect consequences of the defects in intra-Golgi recycling.
Finally, and perhaps most interestingly, we have demonstrated physical and genetic interactions between the Sec34/35p complex and the cytosolic precursor of the COPI coat, coatomer. The COPI vesicle coat was shown to function in both Golgi-to-ER (Letourneur et al., 1994) and in intra-Golgi (Lanoix et al., 1999) retrograde transport of Golgi enzymes and cargo receptors. It may also be involved in transport of anterograde cargo within the Golgi complex (Orci et al., 1997; for review see Pelham and Rothman, 2000). We have demonstrated here that coatomer can be copurified with GST-tagged subunits of the Sec34/35 complex from yeast cells and that coatomer can also be recruited from yeast lysates to recombinant GST-Sec35p purified from E. coli. Interestingly, Sec35p, but not Sec34p was able to interact with both the γ-subunit of coatomer and the GTP-form of ARF1 (unpublished data) in the yeast two-hybrid system, and therefore may serve as the link between coatomer and the Sec34/35p complex. Finally, a deletion of TFI2 gene is shown to be lethal in conjunction with the sec21-1 mutation affecting γ-COP.
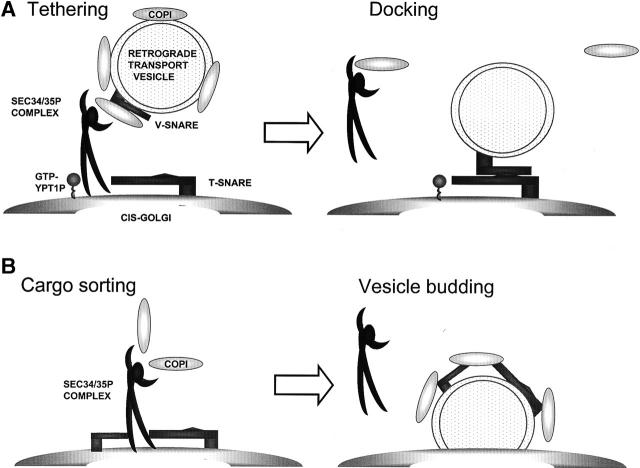
How exactly the Sec34/35p complex contributes to retrograde trafficking remains a question for the future, but we wish to discuss two likely models here. Considering that the Sec34/35p complex binds coatomer as well as the retrograde Golgi SNAREs, it is possible to envision that the Sec34/35p complex is primarily involved in a step between uncoating and docking of the COPI-coated retrograde intra-Golgi vesicles to cis-Golgi membranes (Fig. 10, top). It will be important to determine whether the Sec34/35p complex can bind to both coatomer and Sed5p at the same time or whether the interaction is sequential. Alternatively, the Sec34/35p protein complex could be involved not at a vesicle tethering step of trafficking but during the COPI vesicle formation step (Fig. 10, bottom). The cis-Golgi SNARES Sed5p (Wooding and Pelham, 1998) and Sec22p (Ballensiefen et al., 1998) are known to recycle between Golgi and ER membranes. Interactions between these Golgi SNAREs and the Sec34/35p complex may be necessary for a sorting step in the formation of retrograde vesicles. However, we favor a function for the Sec34/35 complex in vesicle tethering, as (a) both sec34-2 and sec35-1 mutant cells accumulate numerous undocked 50 nm transport vesicles (Wuestehube et al., 1996); (b) structural relationships between Sec34/35 complex and other yeast tethering complexes have been recently uncovered (Pfeffer, 2001; Whyte and Munro, 2001a); and (c) like other vesicle tethering factors, the Sec34/35p complex intimately interacts with the activated, GTP-bound form of the cis-Golgi localized Rab protein Ypt1p.
Figure 10.
Models for Sec34/35p complex function in intra-Golgi protein trafficking. The Sec 34/35 complex may act either in tethering of COPI-coated, retrogradely targeted transport vesicles with Golgi membranes (A), or alternatively, it may be involved in cargo selection into COPI vesicles during budding from Golgi membranes (B). See text for more details.
Materials and methods
Media, plasmids, and strains
YPD, YNB, and sporulation media were prepared as described (Rose et al., 1990). YPGal and YPGlu-5% media contain 1% yeast extract, 2% Bacto-peptone, 2% galactose, or 5% glucose, respectively. Transformation of yeast was performed as described (Elble, 1992); all other standard genetic techniques were done as indicated (Rose et al., 1990). The E. coli strain XL10-Gold (Stratagene), which was used for all molecular genetic manipulations, was grown on standard media (Sambrook et al., 1989) and transformed according to Hanahan (1983).
The plasmids used in this study are listed in Table I and were constructed as follows. To generate pEMBL-GST-Sec34, BamHI–PstI-digested pEMBLyex4 (Baldari et al., 1987) was ligated with the BamHI–PstI fragment of the pSV30 (VanRheenen et al., 1999), which encodes the SEC34 ORF. The TFI1 ORF was amplified by PCR using genomic DNA of Saccharomyces cerevisiae RSY255 as template. Based on the sequence of YGL223c, specific primers were designed so to place BamHI site upstream of the codon for the second amino acid residue of the protein and XhoI site downstream of the stop codon (5′ primer, 5′ cag gat cca tgg atg aag tct ta 3′; 3′ primer, 5′ cgc tcg agt tac tgt tgc ctt aa 3′). A 1,270-bp PCR product was cleaved with BamHI and XhoI, and ligated into a pEMBLyex4 digested by BamHI and SalI, yielding a C-terminal fusion to GST. The construct expressing the GST-TFI2 was constructed in a similar manner, also placing BamHI site upstream of the codon for the second amino acid residue of the TFI2 and a SalI site downstream of the stop codon (5′ primer, 5′ cgg gat cca tgg att tcg ttg ta3′; 3′ primer, 5′ cgg tcg act cag tga tca ata cc 3′). A 2,536-bp PCR product was cleaved with BamHI and SalI and cloned into pBluescript digested with BamHI and XhoI. The YNL041c ORF was subcloned into the pEMBLyex4 as BamHI fragment. The S. cerevisiae strains used in this paper are listed in Table II.
Table I. Plasmids used in this work.
| Plasmid | Description | Source |
|---|---|---|
| pYGST-TFI1 | P Gal -GST-TFI1 in pEMBLyex4 (2 μ, URA3) | This study |
| pYGST-TFI2 | P Gal -GST-TFI2 in pEMBLyex4 (2 μ, URA3) | This study |
| pYGST-SEC34 | P Gal -GST-SEC34 in pEMBLyex4 (2 μ, URA3) | This study |
| pYGST-TFI3 | P cup1 -GST-TFI3 in pYEX4t-1mod | Invitrogen |
| pNB167 | YPT1, 2 μ, URA3 | S. Ferro-Novicka |
| pWB-Acycα | P CYC1 -SEC22-myc-α, CEN, URA3 | H.D. Schmitt |
| pSV29 | GST-SEC35 in pGEX4T | M.G. Watersb |
| pGST-Sed5 | GST-SED5(1-324) in pGEX2T | M.G. Waters |
| pGAL10-GST-YPT1 | GST-YPT1 in pEMBLyex4 (2 μ, URA) | N. Segevc |
| pSV30 | His 6 -SEC34 in pQE30 | M.G. Waters |
Yale University, New Haven, CT.
Princeton University, Princeton, NJ.
University of Chicago, Chicago, IL.
Table II. S. cerevisiae strains used in this work.
| Strain | Genotype | Source |
|---|---|---|
| LY173 | MATa pep4-HIS3 prb::HisG prc::HisG ura1-3 leu2-3,-112 trp1-1 ade2-1 can1-100 pEMBLyex4 (2 μ, PGAL-GST, URA3) | This study |
| LY186 | MATa pep4-HIS3 prb::HisG prc::HisG leu2-3,-112 trp1-1 ade2-1 can1-100 sec35Δ::SEC35-TAP | This study |
| LY208 | MATa ura3-52 leu2-3,-112 trp1-1 sec34Δ::LEU2 pYGST-SEC34 (2 μ, P GAL -GST-SEC34 URA3) | This study |
| LY236 | MATa/α ura3/ura3 leu2/leu2 his3/his3 lys2/LYS2 met15/MET15 tfi2Δ::Gm/tfi2Δ::Gm | Invitrogen |
| LY237 | MATa/α ura3/ura3 leu2/leu2 his3/his3 lys2/LYS2 met15/MET15 tfi1Δ::Gm/ tfi1Δ::Gm | Invitrogen |
| LY238 | MATa/α ura3/ura3 leu2/leu2 his3/his3 lys2/LYS2 met15/MET15 tfi3Δ::Gm/TFI3 | Invitrogen |
| LY241 | MATa ura3 leu2 his3 lys2 met15 tfi1Δ::Gm | Invitrogen |
| LY291 | MAT ura3-52 leu2-3,-112 trp1-1 sec34D::LEU2 pNB167 (2 μ, YPT1, URA3) | This study |
| LY293 | MATα ura3 leu2 his3 tfi3D::Gm pYGST-TFI3 (2 μ, P CUP -GST-TFI3, URA3) | This study |
| LY297 | MATα ura3 leu2 his3 lys2 met15 (2 μ, YPT1, URA3) | This study |
| LY314 | MATα ura3 leu2 his3 tfi3D::Gm pNB167 (2 μ, YPT1, URA3) | This study |
| LY330 | MATa/α ura3/ura3 leu2/leu2 his3/his3 lys2/LYS2 met15/MET15 tfi1Δ::Gm/ tfi1Δ::Gm pYGST-TFI1 (2 μ, P GAL -GST-TFI1, URA3) | This study |
| LY344 | MATa/α ura3/ura3 leu2/leu2 his3/his3 lys2/LYS2 met15/MET15 tfi2Δ::Gm/ tfi2Δ::Gm pYGST-TFI2 (2 μ, P GAL -GST-TFI2, URA3) | This study |
| LY367 | MATa pep4-HIS3 prb::HisG prc::HisG leu2-3,-112 trp1-1 ade2-1 can1-100 tfi3Δ::TFI3-TAP | This study |
| RSY255 | MATα ura 3-52 leu 2-3, 112 | R. Schekman |
| RSY277 | MATα sec21-1 ura3-52 | R. Schekman |
| RSY1157 | MATa pep4-HIS3 prb::HisG prc::HisG ura1-3 leu2-3,-112 trp1-1 ade2-1 can1-100 | R. Schekman |
| GWY93 | MATα sec35-1 ura3-52 leu2-3,-112 | M.G. Waters |
| GWY95 | MATα sec34-2 ura3-52 leu2-3,-112 lys2-801 | M.G. Waters |
| GWY234 | MATα sed5-1 ura3-52 leu2-3,-112 lys2-801 ade2-1 trp1-1 | M.G. Waters |
| EGY48 | MATα, his3, trp1, ura3-52, leu2::pLEU2-LEXAop3 | E. Golemisa |
| GWY235 | MATα, sed5-1, ura3-52, leu2-3, -112, Lys2-801, trp1-1 | M.G. Waters |
Fox Chase Cancer Center, Philadelphia, PA.
TAP-tagging and Sec34/Sec35p complex purification
SEC35 and TFI3 were tagged at their C termini and integrated into the genome of protease deficient strain RSY1157 by transformation with PCR product from the template plasmid pBS1539, encoding calmodulin-binding peptide, a TEV protease cleavage site, the protein A tag, and the URA3 marker (Rigaut et al., 1999). Integration of this product results in fusion to the SEC35 ORF of a 20.5-kD tag.
For large-scale isolation of Sec35-TAP, 10 liter cultures were grown to an OD600 of ∼3.0, harvested, and washed with distilled water. The pellet was resuspended in an equal volume of ice-cold lysis buffer (LyB, 20 mM Tris-HCl, pH 8.0, 0.1 M NaCl, 5% glycerol, protease inhibitors cocktail; Roche). An equal volume of glass beads was added and the mixture vortexed three times for 1 min each, with intervals on ice. Unbroken cells were separated by centrifugation at 2,000 g for 5 min. Cell membranes were separated by high-speed centrifugation at 150,000 g for 1 h and the supernatant (S150, 10 mg protein/ml) was used for complex purification. (NH4)2SO4 was added to 35% saturation, dissolved, and the solution was stirred for 60 min in the cold room. The (NH4)2SO4 precipitate was collected by centrifugation at 20,000 g for 30 min, resuspended in 10 ml of LyB with 0.5 mM EDTA and 0.1% NP-40 without protease inhibitors (LyBN), and the insoluble material was removed by centrifugation (20,000 g for 30 min). 10 ml of soluble material (∼5 mg protein/ml) was mixed with 0.2 ml of IgG-agarose (Sigma-Aldrich) and incubated for 2 h (4°C, rotation). Beads were then washed three times with 20 ml LyBN buffer and transferred to a 5-ml plastic column (Pierce Chemical Co.). Bound complex was eluted by adding five bead volumes of LyBN containing 100 U of TEV protease (Life Technologies), incubating at 10°C overnight, and collecting the (TEV) eluate by gravity flow. CaCl2 was added to 2 mM, and the TEV eluate was mixed with 0.2 ml of the calmodulin-agarose beads (Sigma-Aldrich). Beads were incubated for 1 h in the cold room, washed four times with LyB, and bound material was eluted with 0.5 ml of 2-mM EGTA in LyB buffer. Proteins were precipitated with DOC/TCA method (Bensadoun and Weinstein, 1976) and separated on 7.5% SDS-PAGE. Coomassie R-250–stained bands were excised, digested with trypsin, and subjected to MALDI-TOF spectroscopy (Shevchenko et al., 1996).
Purification of GST-tagged proteins from the yeast, electrophoresis, and immunoblot analysis
To assay for protein–protein interactions in vivo, GST-tagged proteins and associated proteins were purified from appropriate yeast strains. 5 ml of an overnight culture grown in selective medium containing 1% glucose was used to inoculate 100 ml fresh selective medium containing 2% galactose to induce the expression of the GST fusion proteins. Cells were grown at 30°C to midlogarithmic phase (OD600 = 1.5) and were washed with ice-cold water. 1 vol of glass beads and 1 vol of the buffer (20 mM Hepes, pH 7.4, with protease inhibitors cocktail; Roche) was added. The material was vortexed six times for 1 min each, with intervals on ice. Unbroken cells were separated by centrifugation at 2,000 g for 5 min. P150 fraction was obtained by centrifugation of S2 lysate at 150,000 g for 1 h at 4°C in Beckman TLA55 rotor. The pellet was suspended in extraction buffer (20 mM Hepes, pH 7.4, 1% CHAPS, 0.15 M NaCl) and incubated on ice for 20 min. This membrane extract (1 ml) was cleared by centrifugation at 20,000 g for 10 min at 4°C and mixed with 50 μl prewashed glutathione–Sepharose beads (Amersham Pharmacia Biotech). After a 30-min incubation (4°C, rotation), the beads were washed four times with 20 vol of washing buffer (20 mM Hepes, pH 7.4, 0.1% CHAPS, 0.15 mM NaCl). Bound proteins were eluted using one bead volume (50 μl) of 20 mM reduced glutathione, pH 7.5. Samples for immunoblotting were separated by SDS-PAGE (12% acrylamide, except where noted), electrotransferred to nitrocellulose, and probed with the appropriate primary antibodies according to standard protocols (Harlow and Lane, 1988). Monoclonal antibodies against the c-myc epitope (9E10) were used at a dilution of 1:2,000. Affinity-purified antibodies against Sec34p, Sec35p (VanRheenen et al., 1999), Sed5p, Sec22p, Ypt1p, GST (Lupashin and Waters, 1997), Bet1p, and Bos1p (Sapperstein et al., 1996), and sera against gp400/hsp150 (Lupashin et al., 1992), Gos1p, Ykt6p, CPY, invertase (Lupashin et al., 1997), Kar2p, coatomer (Duden et al., 1994), Sec21p, and Sec61p, a gift from R. Schekman (University of California at Berkeley, Berkeley, CA), α-factor, a gift from H.-D.Schmitt (Max-Planck Institute, Goettingen, Germany), Sso2p, Snc1, a gift from P. Brennwald (Weil Medical College, Ithaca, NY), were all used at a dilution of 1:2,000. Horseradish peroxidase–conjugated goat anti–rabbit or goat anti–mouse secondary antibody (Jackson ImmunoResearch Laboratories) was used at a dilution of 1:3,000. Immunoblots were developed with the Chemiluminescence Reagent Plus kit (NEN Life Science Products).
In vitro protein–protein interactions
His6-Sec34p (VanRheenen et al., 1999), GST-Sed5p (Lupashin and Waters, 1997), and GST-Sso1p (Nicholson et al., 1998) were expressed in E. coli XL10-Gold cells and purified as described previously (Lupashin and Waters, 1997; Lupashin et al., 1997). GST-Ypt1p was expressed in RSY255 yeast cells transformed with pEMBLyex4-YPT1 plasmid. Plasmid pRC701 (Calero and Collins, 2002) was used to integrate GST-YPT6 in the genome of RSY225 strain. Fusion protein production was induced by overnight incubation on the selective medium containing 2% galactose. GST-Ypt1 and GST-Ypt6 were purified from digitonin (0.5%) solubilized total cell membranes on the glutathione–Sepharose 4B using manufacturer's protocol (Amersham Pharmacia Biotech). Sec34/35 protein complex was partially purified on human IgG-agarose column from the cytosol of TFI3-TAP–expressing cells (LY335) as described earlier for Sec35-TAP complex purification. The ammonium sulfate precipitation step was omitted.
Purified GST-Sso1 or GST-Sed5p (5 μg each) was incubated for 1 h at 4°C with 15 μl of glutathione–Sepharose 4B beads equilibrated with buffer B (20 mM Hepes-KOH, pH 7.0, 0.15 M KOAc, 5% glycerol). The beads were centrifuged at 1,000 g for 1 min and washed three times with buffer B. Then beads were mixed with His6-Sec34p (5μg) or purified Sec34/35 complex (0.2 μg) and incubated for 3 h at 4°C. The bound proteins were eluted with 20 μl of 10 mM reduced glutathione in 50 mM Tris-Cl (pH 7.5), separated by 10% SDS-PAGE, and stained with Coomassie R-250 or Western blotted with antibodies against Sec34p and Sec35p.
For Ypt-Sec34/35 interaction experiments, GST-Ypt6 or GST-Ypt1p (1 μg each) were incubated with 15 μl of glutathione–Sepharose 4B beads in 0.5 ml of buffer BY (40 mM Hepes-KOH, pH 7.0, 0.1 M KOAc, 1 mM DTT, 5% glycerol) for 30 min at 4°C. EDTA (2 mM final) and nucleotides (GDP or GTP, 1 mM final) were added, and incubation was continued for another 30 min. Mg2+-acetate was added (10 mM final), and incubation was continued for another 30 min. Beads were pelleted by centrifugation (600 g, 1 min) and resuspended in buffer BY supplemented with 2 mM Mg2+-acetate. Purified Sec34/35 complex (0.2 μg) was added and the mixture was incubated for 3 h (4°C, rotation). The beads were washed once with buffer BY, bound proteins were eluted with 20 μl of sample buffer, separated by 10% SDS-PAGE, and Western blotted with antibodies against Sec34p and Sec35p.
Two-hybrid analysis
The LexA two-hybrid System with yeast reporter strain EGY48 and the bait plasmids pEG202 or pEG203 and the prey plasmids pJG4-5 or pJG4-6 (Estojak et al., 1995; Golemis et al., 1996) were used as previously described (Eugster et al., 2000). Prey fusions are under the control of the GAL1 promoter. Briefly, fusion constructs were created by ligating PCR products made using custom primers into vectors above, generating in-frame fusions with LexA (bait vector) or acid blob B42 (prey vector). For full-length fusions of SEC34 and SEC35, the chimeras started at the first ATG codon of the ORFs. Plasmids encoding two-hybrid chimeras of coatomer subunits have been described (Eugster et al., 2000).
Reporter strain EGY48, harboring the LacZ reporter plasmid pSH18-34 (URA3; Golemis et al., 1996), was cotransformed with a bait and a prey fusion construct and transformants selected on –HIS, -TRP, -URA plates. To assess growth on media lacking leucine, four independent transformants were streaked onto plates lacking histidine, tryptophane, uracil, and leucine, containing 2% galactose, 1% raffinose, or 2% glucose, and incubated at 30°C for 3 d. Each experiment was repeated three times.
Glycerol velocity centrifugation
Yeast cytosol (S150, 0.4 ml, 20 mg/ml) was layered onto a 12-ml linear 10–30% glycerol (wt/vol) gradient made in 20 mM Hepes KOH buffer, pH 7.4. The gradients were centrifuged, and fractions were collected and analyzed as previously described (Suvorova et al., 2001).
Acknowledgments
We are very grateful to Kathryn Lilley (MRC Laboratory of Molecular Biology, Cambridge, UK) and Sew Yeu Peak-Chew (University of Cambridge, Cambridge, UK) for protein identification by MALDI mass spectroscopy, and to Martin Dale (Cambridge) for expert technical assistance. We thank Ruth Collins, Richard Kurten, James McNew, Randy Schekman, Hans Dieter Schmitt and Gerry Waters for advice and reagents.
R. Duden is a Senior Research Fellow of The Wellcome Trust (047578), and V. Lupashin is supported by the American Cancer Society Institutional Research Grant (IRG 91-021-08) and by the Arkansas Breast Cancer Research Grant.
Footnotes
Abbreviations used in this paper: CPY, carboxypeptidase Y; TAP, tandem affinity purification; TFI, thirty-five interacting.
References
- Allan, B.B., B.D. Moyer, and W.E. Balch. 2000. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 289:444–448. [DOI] [PubMed] [Google Scholar]
- Andag, U., T. Neumann, and H.D. Schmitt. 2001. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem. 276:39150–39160. [DOI] [PubMed] [Google Scholar]
- Baldari, C., J.A.H. Murray, P. Ghiara, G. Cesareni, and C.L. Galeotii. 1987. A novel leader peptide which allows efficient secretion of a fragment of human interleukin 1β in Saccharomyces cerevisiae. EMBO J. 6:229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballensiefen, W., D. Ossipov, and H.D. Schmitt. 1998. Recycling of the yeast v-SNARE Sec22p involves COPI-proteins and the ER transmembrane proteins Ufe1p and Sec20p. J. Cell Sci. 111:1507–1520. [DOI] [PubMed] [Google Scholar]
- Banfield, D.K., M.J. Lewis, and H.R. Pelham. 1995. A SNARE-like protein required for traffic through the Golgi complex. Nature. 375:806–809. [DOI] [PubMed] [Google Scholar]
- Bensadoun, A., and D. Weinstein. 1976. Assay of proteins in the presence of interfering materials. Anal. Biochem. 70:241–250. [DOI] [PubMed] [Google Scholar]
- Bensen, E.S., B.G. Yeung, and G.S. Payne. 2001. Ric1p and the ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol. Biol. Cell. 12:13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret, E., G. Rigaut, A. Shevchenko, M. Wilm, and B. Seraphin. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19:1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald, P., and P. Novick. 1993. Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature. 362:560–563. [DOI] [PubMed] [Google Scholar]
- Calero, M., and R.N. Collins. 2002. Saccharomyces cerevisiae Pra1p/Yip3p interacts with Yip1p and Rab proteins. Biochem. Biophys. Res. Commun. 290:676–681. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., H.M. McBride, R.D. Burgoyne, and M. Zerial. 1999. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 397:621–625. [DOI] [PubMed] [Google Scholar]
- Duden, R., M. Hosobuchi, S. Hamamoto, M. Winey, B. Byers, and R. Schekman. 1994. Yeast β-and β′-COP: two coatomer subunits essential for ER-to-Golgi traffic. J. Biol. Chem. 269:24486–24495. [PubMed] [Google Scholar]
- Elble, R. 1992. A simple and efficient procedure for transformation of yeasts. Biotechniques. 13:18–20. [PubMed] [Google Scholar]
- Estojak, J., R. Brent, and E.A. Golemis. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster, A., G. Frigerio, M. Dale, and R. Duden. 2000. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 19:3905–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick, S., and P. Novick. 1993. The role of GTP-binding proteins in transport along the exocytic pathway. Annu. Rev. Cell Biol. 9:575–599. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard, G., S.F. Nothwehr, and T.H. Stevens. 1997. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J. Cell Biol. 137:1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis, E.A., and V. Khazak. 1997. Alternative yeast two-hybrid systems. The interaction trap and interaction mating. Methods Mol. Biol. 63:197–218. [DOI] [PubMed] [Google Scholar]
- Golemis, E.A., J. Gyuris, and R. Brent. 1996. Interaction trap/two-hybrid system to identify interacting proteins. In Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York. 20.1.1.–20.1.28.
- Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 726 pp.
- Hsu, S.C., A.E. Ting, C.D. Hazuka, S. Davanger, J.W. Kenny, Y. Kee, and R.H. Scheller. 1996. The mammalian brain rsec6/8 complex. Neuron. 17:1209–1219. [DOI] [PubMed] [Google Scholar]
- Huffaker, T.C., and P.W. Robbins. 1983. Yeast mutants deficient in protein glycosylation. Proc. Natl. Acad. Sci. USA. 80:7466–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 98:4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd, G., C. Richardson, R. Litt, and N. Segev. 1995. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J. Cell Biol. 131:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S., G. Jedd, R.A. Kahn, A. Franzusoff, F. Bartolini, and N. Segev. 1999. Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics. 152:1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.W., M. Sacher, A. Scarpa, A.M. Quinn, and S. Ferro-Novick. 1999. High-copy suppressor analysis reveals a physical interaction between Sec34p and Sec35p, a protein implicated in vesicle docking. Mol. Biol. Cell. 10:3317–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix, J., J. Ouwendijk, C.C. Lin, A. Stark, H.D. Love, J. Ostermann, and T. Nilsson. 1999. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COPI vesicles. EMBO J. 18:4935–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur, F., E.C. Gaynor, S. Hennecke, C. Démollière, R. Duden, S.D. Emr, H. Riezman, and P. Cosson. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 79:1199–1207. [DOI] [PubMed] [Google Scholar]
- Li, B., and J.R. Warner. 1998. Genetic interaction between YPT6 and YPT1 in Saccharomyces cerevisiae. Yeast. 14:915–922. [DOI] [PubMed] [Google Scholar]
- Lucau-Danila, A., R. Wysocki, T. Roganti, and F. Foury. 2000. Systematic disruption of 456 ORFs in the yeast Saccharomyces cerevisiae. Yeast. 16:547–552. [DOI] [PubMed] [Google Scholar]
- Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. [DOI] [PubMed] [Google Scholar]
- Lupashin, V.V., and M.G. Waters. 1997. t-SNARE activation through transient interaction with a rab-like guanosine triphosphatase. Science. 276:1255–1258. [DOI] [PubMed] [Google Scholar]
- Lupashin, V.V., S.V. Kononova, Y.E. Ratner, A.B. Tsiomenko, and I.S. Kulaev. 1992. Identification of a novel secreted glycoprotein of the yeast Saccharomyces cerevisiae stimulated by heat shock. Yeast. 8:157–169. [DOI] [PubMed] [Google Scholar]
- Lupashin, V.V., S. Hamamoto, and R.W. Schekman. 1996. Biochemical requirements for the targeting and fusion of ER-derived transport vesicles with purified yeast Golgi membranes. J. Cell Biol. 132:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin, V.V., I.D. Pokrovskaya, J.A. McNew, and M.G. Waters. 1997. Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol. Biol. Cell. 8:2659–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, B.J., and H.R. Pelham. 1998. SNAREs and membrane fusion in the Golgi apparatus. Biochim. Biophys. Acta. 1404:9–31. [DOI] [PubMed] [Google Scholar]
- Nicholson, K.L., M. Munson, R.B. Miller, T.J. Filip, R. Fairman, and F.M. Hughson. 1998. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat. Struct. Biol. 5:793–802. [DOI] [PubMed] [Google Scholar]
- Orci, L., M. Stamnes, M. Ravazzola, M. Amherdt, A. Perrelet, T.H. Sollner, and J.E. Rothman. 1997. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell. 90:335–349. [DOI] [PubMed] [Google Scholar]
- Ossipov, D., S. Schroder-Kohne, and H.D. Schmitt. 1999. Yeast ER-Golgi v-SNAREs Bos1p and Bet1p differ in steady-state localization and targeting. J. Cell Sci. 112:4135–4142. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R., and J.E. Rothman. 2000. The debate about transport in the Golgi—two sides of the same coin? Cell. 102:713–719. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. 2001. Vesicle tethering factors united. Mol. Cell. 8:729–730. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R. 1999. Transport-vesicle targeting: tethers before SNAREs. Nat. Cell Biol. 1:E17–E22. [DOI] [PubMed] [Google Scholar]
- Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 24:218–229. [DOI] [PubMed] [Google Scholar]
- Rexach, M.F., and R.W. Schekman. 1991. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J. Cell Biol. 114:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Rose, M., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 198 pp.
- Russo, P., N. Kalkkinen, H. Sareneva, J. Paakkola, and M. Makarow. 1992. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc. Natl. Acad. Sci. USA. 89:3671–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1.1–18.88.
- Sapperstein, S.K., V.V. Lupashin, H.D. Schmitt, and M.G. Waters. 1996. Assembly of the ER to Golgi SNARE complex requires Uso1p. J. Cell Biol. 132:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals, D.F., G. Eitzen, N. Margolis, W.T. Wickner, and A. Price. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 97:9402–9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. [DOI] [PubMed] [Google Scholar]
- Simonsen, A., J.M. Gaullier, A. D'Arrigo, and H. Stenmark. 1999. The Rab5 effector EEA1 interacts directly with syntaxin-6. J. Biol. Chem. 274:28857–28860. [DOI] [PubMed] [Google Scholar]
- Söllner, T., S.W. Whiteheart, M. Brunner, H. Erdjument-Bromage, S. Geromanos, P. Tempst, and J.E. Rothman. 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature. 362:318–324. [DOI] [PubMed] [Google Scholar]
- Spelbrink, R.G., and S.F. Nothwehr. 1999. The yeast GRD20 gene is required for protein sorting in the trans-Golgi network/endosomal system and for polarization of the actin cytoskeleton. Mol. Biol. Cell. 10:4263–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova, E.S., R.C. Kurten, and V.V. Lupashin. 2001. Identification of a human orthologue of Sec34p as a component of the cis-Golgi vesicle tethering machinery. J. Biol. Chem. 276:22810–22818. [DOI] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T.A. Mansfield, R.S. Judson, J.R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627. [DOI] [PubMed] [Google Scholar]
- VanRheenen, S.M., X. Cao, V.V. Lupashin, C. Barlowe, and M.G. Waters. 1998. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J. Cell Biol. 141:1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen, S.M., X. Cao, S.K. Sapperstein, E.C. Chiang, V.V. Lupashin, C. Barlowe, and M.G. Waters. 1999. Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J. Cell Biol. 147:729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mollard, G.F., S.F. Nothwehr, and T.H. Stevens. 1997. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J. Cell Biol. 137:1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, M.G., and S.R. Pfeffer. 1999. Membrane tethering in intracellular transport. Curr. Opin. Cell Biol. 11:453–459. [DOI] [PubMed] [Google Scholar]
- Whyte, J.R., and S. Munro. 2001. a. A yeast homolog of the mammalian mannose 6-phosphate receptors contributes to the sorting of vacuolar hydrolases. Curr. Biol. 11:1074–1078. [DOI] [PubMed] [Google Scholar]
- Whyte, J.R., and S. Munro. 2001. b. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev. Cell. 1:527–537. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., D.D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J.D. Boeke, H. Bussey, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906. [DOI] [PubMed] [Google Scholar]
- Wooding, S., and H.R. Pelham. 1998. The dynamics of Golgi protein traffic visualized in living yeast cells. Mol. Biol. Cell. 9:2667–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuestehube, L.J., R. Duden, A. Eun, S. Hamamoto, P. Korn, R. Ram, and R. Schekman. 1996. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 142:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]