Figure 3. Analysis of light chain CDRs.
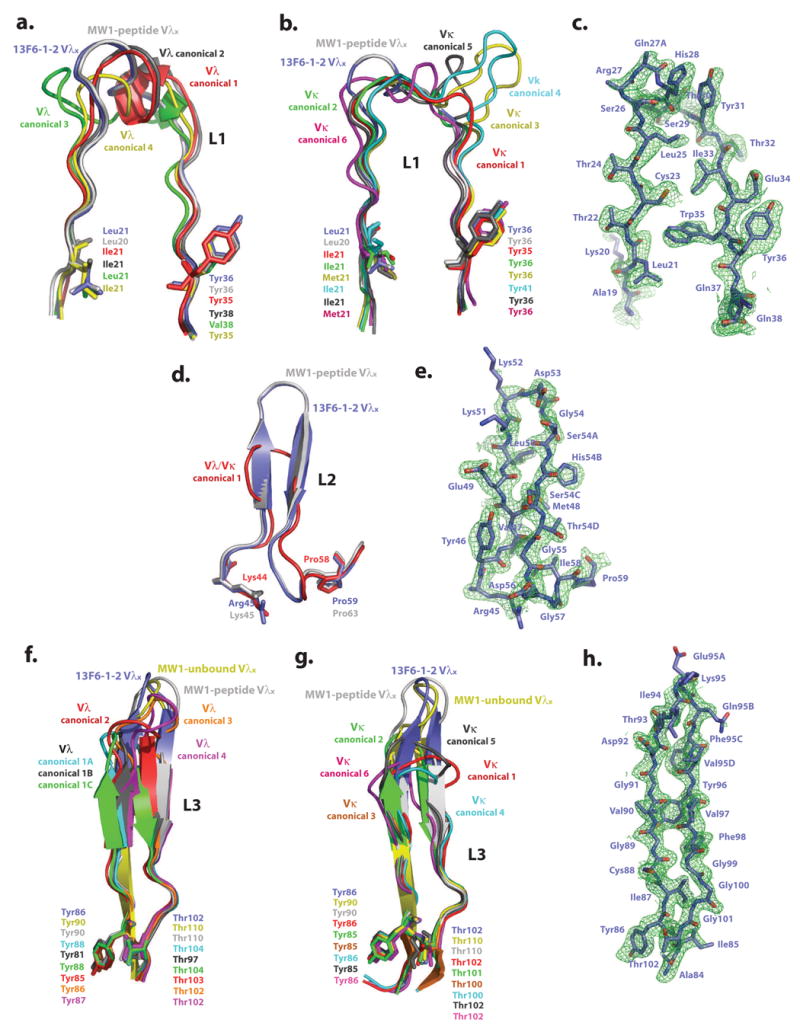
Comparison of Fab 13F6-1-2 and (a) Vλ L1, (b) Vκ L1, (d) Vλ/Vκ L2, (f) Vλ L3, and (g) Vκ L3 CDR canonical conformations. The initial difference σA-weighted Fo-Fc electron density map (green), contoured at 2σ superimposed onto the refined (c) L1, (e) L2, and (h) L3 13F6-1-2 CDR structures. Example structures of CDR canonical conformations are taken from the following antibody structures (P DB accession codes): CDR L1 Vλ canonical conformation 1: 2FB4, 2: 7FAB, 3: 1MFA, 4: 8FAB, MW1-peptide: 2OTU. CDR L1 Vκ canonical conformation 1: 2FBJ, 2: 1VFA, 3: 1HIL, 4: 1FLR, 5: 1GGI, 6: 1FIG, MW1-peptide: 2OTU. CDR L2 Vλ/κ canonical conformation 1: 2FB4, MW1-peptide: 2OTU. CDR L3 Vλ canonical conformation 1A: 1GIG, 1B: 7FAB, 1C: 1MFA, 2: 2FB4, 3: 1Q1J, 4:2B0S, MW1-peptide: 2OTU, MW1-unbound: 2GSG. CDR L3 Vκ canonical conformation 1: 1HIL, 2: 2FBJ, 3: 1BQL, 4: 1DFB, 5: 1BAF, 6: 1EAP, MW1-peptide: 2OTU, MW1-unbound: 2GSG. CDR L1 and L2 of MW1 in the unbound state (PDB code: 2GSG) are not shown in figures 3a, b, and d for figure clarity, as these CDRs are similar in conformation to those observed in the MW1 peptide-bound structure.
