Abstract
A-kinase anchoring proteins (AKAPs) tether the cAMP-dependent protein kinase (PKA) and other signaling enzymes to distinct subcellular organelles. Using the yeast two-hybrid approach, we demonstrate that Rab32, a member of the Ras superfamily of small molecular weight G-proteins, interacts directly with the type II regulatory subunit of PKA. Cellular and biochemical studies confirm that Rab32 functions as an AKAP inside cells. Anchoring determinants for PKA have been mapped to sites within the conserved α5 helix that is common to all Rab family members. Subcellular fractionation and immunofluorescent approaches indicate that Rab32 and a proportion of the cellular PKA pool are associated with mitochondria. Transient transfection of a GTP binding–deficient mutant of Rab32 promotes aberrant accumulation of mitochondria at the microtubule organizing center. Further analysis of this mutant indicates that disruption of the microtubule cytoskeleton results in aberrantly elongated mitochondria. This implicates Rab32 as a participant in synchronization of mitochondrial fission. Thus, Rab32 is a dual function protein that participates in both mitochondrial anchoring of PKA and mitochondrial dynamics.
Keywords: protein kinase; cAMP; anchoring protein; Rab protein; mitochondria
Introduction
The transmission of environmental cues to precise sites within the cell frequently involves the assembly of multiprotein signaling complexes (Pawson, 1995; Hunter, 2000; Jordan et al., 2000). These transduction units, which often include effector proteins, signal transduction enzymes, and their substrate proteins, are maintained by scaffolding or anchoring proteins (Smith and Scott, 2002). These “signal-organizing” molecules ensure that their complement of anchored enzymes are optimally positioned to receive activation signals and are placed in close proximity to their substrates. One example is the A-kinase anchoring proteins (AKAPs)* that sequester the cAMP-dependent protein kinase (PKA) and other signaling enzymes at a variety of intracellular sites (Colledge and Scott, 1999; Feliciello et al., 2001).
Although a principle function of many kinase anchoring proteins is to orchestrate protein phosphorylation events, it is now evident that they participate in a wider range of signaling events. For example, several anchoring proteins have been implicated in the recruitment and localization of Ras family small molecular weight GTPases. AKAP-Lbc binds PKA and functions as a Rho-selective guanine nucleotide exchange factor (GEF) to induce actin stress fiber formation in fibroblasts (Diviani et al., 2001). Scar/WAVE-1 assembles a signaling complex that includes PKA, the Abelson tyrosine kinase, Rac-1, and the Arp2/3 complex to coordinate lamellapodial extension at the leading edge of motile cells (Westphal et al., 2000). Anchoring proteins that bind to other second messenger–regulated protein kinases apparently perform analogous functions. The mammalian orthologue of the Caenorhabditis elegans cell polarity gene, mPAR-6, interacts with either Cdc42 or Rac1 and an atypical PKC to establish cell shape and polarity in the developing embryo (Etemad-Moghadam et al., 1995; Kuchinke et al., 1998; Joberty et al., 2000; Lin et al., 2000; Qiu et al., 2000). Collectively, these findings highlight a secondary role for kinase anchoring proteins in the coordination of small G-protein location and signaling to downstream effectors.
Here we report that Rab32, a member of the Rab subfamily of Ras small molecular weight G-proteins, functions as an AKAP in vivo. Most Rab proteins have been implicated in membrane dynamics, where they coordinate the assembly of protein networks that regulate membrane fusion/fission, exocytosis, and cytoskeletal trafficking (Takai et al., 2001). These events require the Rab proteins to cycle from a cytoplasmic-localized GDP-bound “off” state to a membrane-localized GTP-bound “on” state (Novick and Zerial, 1997). Accordingly, Rab proteins have been found at various intracellular sites including the endoplasmic reticulum, the Golgi apparatus, endosomes, intracellular vesicles, and the plasma membrane (Takai et al., 2001). Our cell-based studies demonstrate that Rab32 is associated with the mitochondria through potential lipid modification of two COOH-terminal cysteines. Furthermore, ectopic expression of a GTP binding–defective mutant of Rab32 perturbs mitochondrial distribution, suggesting a specific role for this Rab protein at mitochondria.
Results
Identification and characterization of Rab32 as a type II regulatory subunit (RII) binding protein
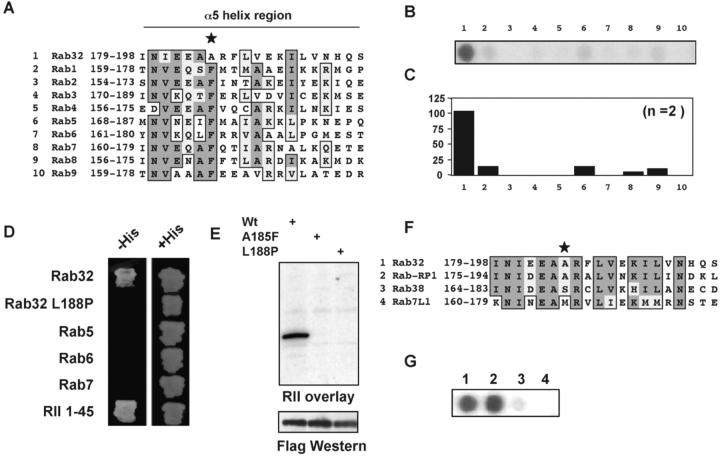
Biochemical and structural analyses have indicated that the first 45 amino acids of the type II regulatory subunit (RII) comprise the primary determinants for interacting with AKAPs (Hausken et al., 1994; Newlon et al., 1999). To identify new AKAPs, the first 45 amino acids of the murine RIIα were used as “bait” to screen a human brain cDNA library using the yeast two-hybrid assay. 50 positive colonies were identified from 2 × 106 cotransformants, as assessed by activation of both the HIS3 and LacZ genes. 40 of these positive colonies encoded previously identified AKAPs, including Yotiao, AKAP220, and S-AKAP84 (Fig. 1 A). One positive colony encoded residues 106–225 of Rab32 that, based on primary amino acid similarity and domain architecture, is believed to be a small molecular weight G-protein of the Rab family (GenBank/EMBL/DDBJ accession no. NP_006825) (Bao et al., 2002). Independent confirmation of this result was provided when the bacterially expressed Rab32 106–225 fragment bound recombinant [32P]RIIα, as assessed by a solid-phase binding assay (Fig. 1 B).
Figure 1.
Identification of Rab32 as a putative AKAP. Murine RIIα 1–45/LexA fusion was used as bait to screen a human brain cDNA library using the yeast two-hybrid assay. (A) Detection of positive colonies expressing Rab32 and other AKAP fragments were detected by β-galactosidase. The name of each AKAP is indicated. (B) Control and bacterial extracts expressing the Rab32 106–225 fragment (indicated above each lane) were separated on SDS-PAGE (4–15%) and electrotransferred to nitrocellulose. RII binding was assessed by overlay using 32P-labeled RIIα and detected by autoradiography. Molecular weight markers are indicated.
The PKA anchoring site on AKAPs includes a region of 14–18 residues that forms an amphipathic helix (Carr et al., 1991; Newlon et al., 2001). To define this region within Rab32, an array of overlapping 20-mer peptides (offset by three amino acids) covering residues 151–225 were synthesized on a membrane support using an AutoSpot robot (Fig. 2 A). A single peptide corresponding to amino acids 178–197 (NINIEEAARFLVEKILVNHQ) strongly bound RIIα, as assessed by the solid-phase binding assay (Fig. 2 B). Computer predictions of its secondary structure suggest that residues 178–197 form an amphipathic helix (Fig. 2 C). All Ras family members share a similar structure with an arrangement of six central strands of β-sheet and five α-helical regions. Structural analysis of Rab3a, a protein with 71% identity to Rab32, suggests that the RII binding region falls within the conserved α5 helix (Ostermeier and Brunger, 1999). Modeling studies using the coordinates from the Rab3a structure suggest that the α5 helix of these proteins are located on an exposed surface of the molecule and would be available for docking to RII (Fig. 2 D). Experimental confirmation of these observations was provided when the Rab32 175–205 peptide blocked RIIα interaction with a full-length Rab32 GST fusion protein (Fig. 2 E, middle). A control peptide, where leucine 188 was replaced with proline to disrupt secondary structure, was unable to block RII interaction (Fig. 2 E, right). These data imply that RII interacts with Rab32 through an amphipathic helix formed by residues 178–197 of the protein. Solution binding assays confirmed that recombinant Rab32 bound [32P]RIIα, whereas the Rab32L188P mutant did not (Fig. 2 F). Together, these results show that Rab32 can interact with full-length RII in vitro and this interaction occurs through a predicted amphipathic helix in a manner similar to other AKAPs.
Figure 2.
Mapping the RII binding domain of Rab32. (A) Schematic representation of the mapping strategy to identify the RII binding domain of Rab32. A family of 20-mer peptides (each offset by three residues) spanning the region 153–224 of Rab32 were synthesized and immobilized to a membrane support. (B) Solid-phase binding of RII was assessed by the RII overlay procedure. The sequence of the RII binding peptide is indicated using the one letter amino acid code (*). (C) Ribbon diagram of the Rab3a backbone based upon coordinates provided by Ostermeier and Brunger (1999). Top (left) and side (right) views are presented. The α5 helix is marked (red). (D) Helical wheel alignment of residues 181–193 of Rab32. Hydrophobic (white circles) and hydrophilic (yellow circles) residues are indicated. (E) Purified recombinant GST–Rab32 fusion protein (2 μg) was separated by SDS-PAGE and electrotransferred to nitrocellulose. RII overlays were performed in the absence of competitor peptide (left), in the presence of 10 μM Rab32 175–205 peptide (middle), or in the presence of 10 μM Rab32 175–205 L188P peptide (right). Molecular weight markers are indicated. (F) Solution binding of recombinant RII and Rab32. [32P]RIIα was incubated with either GST–Rab32 or GST–Rab32L188P mutant followed by glutathione-Sepharose purification of the complex. RII binding was assessed by measuring the counts per minute (cpm) corresponding to [32P]RIIα bound to Rab32 (top). Equal amounts of GST fusion proteins were used in these experiments (bottom).
We next sought to determine if Rab32 is an AKAP in vivo. Flag-tagged versions of wild-type Rab32 and Rab32L188P mutant were expressed in HEK-293 cells. Immune complexes were isolated, and coprecipitation of PKA activity was measured by the filter paper assay (Corbin and Reimann, 1974). PKA activity was enriched 7.1 ± 2–fold (n = 3) in Rab32 immune complexes, whereas there was no enhancement of kinase activity in the Rab32L188P mutant immunoprecipitates (Fig. 3 A). Control experiments confirmed that this was specific PKA activity, because the PKI (5–24) peptide, a specific inhibitor of the kinase, blocked all activity (Fig. 3 A). In parallel studies, only wild-type Rab32 bound RII in the Flag immunoprecipitates, as assessed by the RII overlay (Fig. 3 B), suggesting that Rab32 is the only AKAP responsible for the PKA activity. Thus, Rab32 associates with endogenous PKA holoenzyme inside mammalian cells.
Figure 3.
Rab32 can interact with the holoenzyme of PKA in mammalian cells. HEK-293 cells were transiently transfected with Flag–Rab32 or Flag–Rab32L188P cDNA. Triton X-100–soluble extracts were immunoprecipitated with Sepharose-conjugated antiFlag antibody. (A) Copurification of the PKA holoenzyme was measured by assaying for PKA catalytic subunit activity stimulated by exogenous cAMP. PKA activity was measured as pmol/min/mg of 32P incorporated into the PKA substrate kemptide using a filter paper binding assay (Corbin and Reimann, 1974). Specific PKA activity was blocked by 10 μM of the inhibitor PKI (5–24) peptide. (B) AntiFlag immunoprecipitates were separated on SDS-PAGE after the catalytic subunit had been eluted from the complex with exogenous cAMP and electrotransferred to nitrocellulose. RII overlay assays (top) were performed to determine if Rab32 was the only AKAP present in these fractions. Control experiments confirmed that equal levels of protein were immunoprecipitated in these experiments (bottom). A representative example of three independent experiments is shown.
Rab32 and closely related orthologues are the only Rab family members that function as AKAPs
Each of the 60 genes that encode Rab proteins in the human genome is highly conserved in the α5 helix (Bock et al., 2001). Therefore, peptides corresponding to the α5 helix region from nine other Rab proteins (sequences are presented in Fig. 4 A as a clustal alignment) were synthesized by the AutoSpot robot to determine if other Rab family members could interact with RII. Solid-phase binding to RII was only detected for the Rab32 peptide (Fig. 4, B and C). Likewise only full-length Rab32 bound RII, as assessed by the two-hybrid assay (Fig. 4 D). Structural studies on several AKAPs have established that alanines at position 6 of the PKA binding helix are critical anchoring determinants (Newlon et al., 2001). The methyl group of the alanine side chain inserts into a hydrophobic pocket formed by the RII dimer. Rab32 has an alanine at this position, whereas most Rab family members have a phenylalanine at this site in the α5 helix (Fig. 4 A, star). To test the role of Ala 185 as an anchoring determinant, we introduced a phenylalanine by site-directed mutagenesis. The Rab32A185F mutant protein was unable to bind RII in the overlay procedure (Fig. 4 E), and an A185F-substituted peptide was also unable to interact with RIIα (unpublished data). Database searches identified a Drosophila orthologue, Rab-RP1, that contained an alanine in a position analogous to residue 185 of Rab32. Peptide array analysis confirmed that Rab-RP1 bound RII in the overlay assay, whereas two closely related Rab proteins lacking the conserved phenylalanine (Rab38 and Rab7L1) did not bind RII (Fig. 4, F and G). Collectively, these data suggest that Rab32 and orthologues are AKAPs.
Figure 4.
Only Rab32 and RabRP-1 contain determinants for PKA anchoring. (A) The sequences of Rab32 and nine other Rab proteins are aligned within the α5 helix region. The first and last residues of each sequence and the name of each Rab protein are indicated. Boxed and shaded regions depict sequence identity and similarity. Star indicates the conserved phenylalanine in most Rab proteins. (B) A solid-phase peptide array of these Rab sequences was screened for RII binding by the overlay assay. Binding of [32P]RIIα was detected by autoradiography. (C) Quantitation of RII binding was assessed by densitometry. Arbitrary units were normalized to one, indicating the highest level of RII binding. The data presented are an amalgamation of two independent experiments. (D) Yeast two-hybrid analysis was used to determine if RII interacts with selected, full-length Rab family members. Two-hybrid crosses of RIIα 1–45 fragment with full-length cDNAs for Rab32, Rab32L188P, Rab5, Rab6, or Rab7 fused to the LexA DNA binding domain. Interactions were assayed by growth on minimal media plates in the absence of histidine (left). As a toxicity control, all cotransformed yeast strains were able to grow in the presence of histidine (right). (E) HEK-293 cell extracts expressing Flag-tagged Rab32 and mutants (indicated above each lane) were separated by SDS-PAGE on 4–15% gels and electrotransferred to nitrocellulose. Binding of [32P]RIIα was assessed by the overlay assay. (Top) RII binding was detected by autoradiography. Equal loading of recombinant Rab32 proteins was confirmed by Western blot using antiFlag antibodies. (F) Sequence alignment of Rab32 and its most closely related family members. The first and last residues of each sequence and the name of each Rab protein are indicated. Boxed and shaded regions depict sequence similarity and identity. Star indicates the position of alanine 185 in the Rab32 sequence. (G) A solid-phase peptide array of these closely related Rab sequences was screened for RII binding by the overlay assay. Binding of [32P]RIIα was detected by autoradiography.
Rab32 is associated with mitochondria
The tissue distribution of Rab32 mRNA was determined by Northern blot using a specific 113-bp probe. An mRNA species of 1.4 kb was detected prominently in the liver and, to a lesser extent, in other tissues (Fig. 5 A). RNA dot blot analysis detected high levels of Rab32 mRNA in bone marrow, testis, colon, and fetal lung (unpublished data). This latter observation prompted us to examine the subcellular distribution of the Rab32 protein in the human fetal lung fibroblast cell line WI-38. As a prelude to these studies, antipeptide antibodies were raised against residues 202–220 of Rab32 and affinity purified against the recombinant protein. A single protein band of 27 kD was detected in cell extracts as assessed by immunoblot (Fig. 5 B, left). Preincubation with the antigenic peptide blocked antibody recognition of this 27-kD protein (unpublished data). Control experiments using the preimmune serum were negative (Fig. 5 B, right).
Figure 5.
The cellular and subcellular distribution of Rab32. The tissue distribution of Rab32 mRNA was assessed by Northern blot analysis. (A) A human multi-tissue (tissue sources indicated above each lane) was screened using a 113-bp cDNA probe that corresponds to the COOH-terminal hypervariable region of Rab32. Hybridization was detected by autoradiography. The sizes of DNA markers are indicated. (B) Triton X-100–soluble extracts from the WI-38 human fetal lung fibroblasts were immunoblotted with affinity-purified anti-Rab32 antibody (left) or preimmune sera (right). Signals were detected by chemiluminescence. Molecular weight standards are indicated. Confocal immunofluorescence microscopy of WI-38 fibroblasts triple labeled with polyclonal antibodies against Rab32 (C, green), cell-permeable dye MitoTracker RedTM (D, red), and a monoclonal antibody against α-tubulin (E, blue). A merged image (F) indicates the cellular distribution of all three signals. (G) Subcellular fractionation of WI-38 cells was performed according to the Materials and methods. Protein (20 μg each) from whole cell (W), nuclear (P1), and mitochondria-enriched (P2) fractions were subjected to SDS-PAGE and electrotransferred to nitrocellulose. Membranes were immunoblotted using affinity-purified polyclonal anti-Rab32 antibody (top) and a monoclonal anticytochrome oxidase subunit I (bottom) as a marker for mitochondria. Immunocytochemistry and confocal analysis were used to demonstrate the subcellular location of RII (H) and Rab32 (I). A merged image shows a significant overlap of the signals (J).
Immunofluorescent analysis of Rab32 in WI-38 fibroblasts revealed a staining pattern (Fig. 5 C) that overlapped with the mitochondrial marker MitoTracker RedTM (Fig. 5 D) but was distinct from tubulin (Fig. 5 E). This is most clearly demonstrated in a composite image (Fig. 5 F). In support of this observation, Rab32 was enriched in the P2 fraction (Fig. 5 G, top) when WI-38 cell extracts were subjected to subcellular fractionation. The partitioning of Rab32 correlated with the detection of the mitochondrial marker cytochrome oxidase subunit 1 (Fig. 5 G, bottom). Immunofluorescence data also suggested that a proportion of cellular RII signal is present at mitochondria (Fig. 5 H) and overlaps with the Rab32 signal (Fig. 5, I and J). Collectively, these results suggest that Rab32 and RII associate with mitochondria.
Mitochondria are arranged around the microtubule organizing center and utilize microtubules for their movement. However, Rab32 association with mitochondria is not dependent on the integrity of the microtubule cytoskeleton (Fig. 6 , A–H). Cos7 cells were treated with the microtubule depolymerizing agent nocodazole and the location of Rab32, mitochondria, and microtubules were visualized by immunofluorescence detection (Fig. 6, E–H). As expected, nocodazole treatment disrupted the microtubule network (Fig. 6 G) and promoted a redistribution of mitochondria (Fig. 6, F and H), when compared with control cells (Fig. 6, B–D). Importantly, Rab32 was retained on the mitochondria after drug treatment (Fig. 6, E and H), indicating that it specifically associates with this organelle.
Figure 6.

Mitochondrial targeting of Rab32. Immunofluorescence analysis of Rab 32 location upon depolymerization of microtubules in Cos7 cells. Control (A–D) or Cos7 cells treated with 5 μM nocodazole for 30 min (E–H) were fixed and stained. Immunocytochemistry was performed using anti-Rab32 (A and E, green), MitoTracker RedTM (B and F, red), and a monoclonal antibody against α-tubulin (C and G, blue). Composite images are shown for each sample (D and H). (I) Schematic representation of a nucleotide binding site, GTPase domain, and sites of putative lipid modification are indicated. Arrows indicate an amino acid substitution. (J) Flag-tagged wild-type or Rab32ΔCC mutant were expressed in Cos7 cells. The subcellular distribution of recombinant Rab32ΔCC (J) or Rab32 (M) were analyzed using confocal fluorescence microscopy with an antiFlag monoclonal antibody and Texas red–conjugated secondary (J and M). Mitochondria were visualized by using Mito–GFP targeting construct (K and N). Merged images are presented (L and O).
Mitochondrial targeting sequences have been identified in a number of proteins, including other AKAPs (Lin et al., 1995; Huang et al., 1999), yet no such sequences were evident in Rab32. Previous studies have proposed that lipid modification of two COOH-terminal cysteines may participate in the membrane targeting of Rab proteins (Takai et al., 2001). We generated a Flag-tagged form of Rab32 that lacked the pair of COOH-terminal cysteine residues (Fig. 6 J). The Rab32ΔCC mutant did not associate with mitochondria when expressed in Cos7 cells (Fig. 6, K and L). Mitochondria were detected by coexpression of a GFP fusion protein that contained a mitochondrial targeting signal (Fig. 6, K and N). Control experiments demonstrated that wild-type Rab32 codistributed with the Mito–GFP tag when both constructs were transfected into Cos7 cells (Fig. 6, M–O). Additional controls demonstrated that the Rab32L188P mutant, which is unable to anchor PKA, was correctly targeted to mitochondria (unpublished data). These findings suggest that COOH-terminal lipid modification of Rab32 contributes to mitochondrial targeting, most likely to the outer mitochondrial membrane.
Biochemical and cellular analyses of Rab32
Rab32 has four conserved guanine nucleotide binding regions including a possible GTPase domain (Fig. 7 A). Using an in vitro binding assay we found that [3H]GDP dissociated from Rab32 with a t 1/2 of 7.5 min (n = 3; Fig. 7 B). Using a similar assay, GTP binding was measured with a t 1/2 of 12 min (n = 3; Fig. 7 C). These studies show, as predicted, that Rab32 is a guanine nucleotide binding protein. On the basis of homology to other Ras family members, we predicted that mutation of threonine 39, which lies within the GTP binding domain, would abolish interaction with this nucleotide (Fig. 7 A). As expected, the Rab32T39N mutant bound GTP to a 12-fold lesser extent when compared with the wild-type protein (Fig. 7 D). Equal amounts of both Rab32 forms were used in these assays (Fig. 7 D, insert). We next sought to determine if RII preferentially interacts with Rab32 in a nucleotide-dependent manner. Rab32 was preloaded with either GDP or the nonhydrolyzable GTP analogue, GTPγS. Rab32 could associate with RII whether it was loaded with GDP or GTPγS (Fig. 7 E). Yeast two-hybrid assay confirmed this result, as the Rab32T39N mutant and Rab32Q85L, a potential GTPase inactive form, interacted with RII (Fig. 7 F). These data show that Rab32 interacts with RII in a nucleotide-independent state.
Figure 7.
Nucleotide binding properties and GTPase activity of Rab32. Bacterially purified GST fusion proteins were assayed for guanine nucleotide binding. (A) Schematic diagram depicting a mutation in the nucleotide binding domain of Rab32. A nucleotide binding site, GTPase domain, and sites of putative lipid modifications are indicated. (B) GST–Rab32 (2 μg) was preloaded with 2 μCi of [3H]GDP. GDP off rate is presented as percent GDP bound to Rab32 over time. (C) Nucleotide-depleted Rab32 was incubated with [γ32P]GTP. The GTP on rate for GST–Rab32 was measured and is presented as the percentage of GTP bound over time. Amalgamated data from three experiments are presented. (D) Biochemical characterization of guanine nucleotide binding to recombinant wild-type GST–Rab32 and GST–Rab32T39N mutant. [3H]GDP binding (gray bars) and [γ32P]GTP binding (black bars) to Rab32 and Rab32T39N are indicated. Results are presented as the percent nucleotide bound normalized to 1. 2 μg of GST recombinant proteins were used and shown in the inset. A representative experiment from three independent analyses is presented. (E) GST pulldowns of GDP-loaded Rab32 or GTPγS-loaded Rab32 in the presence of recombinant RII (1 μg). Proteins were separated by SDS-PAGE and transferred to nitrocellulose. (Top) Coprecipitation of RII was detected by Western blot using an anti-RII monoclonal antibody. (Bottom) Control experiments showing equal loading of proteins. (F) Yeast coexpressing pLexA Rab32 mutants and pACT2 RII 1–45 were assayed for growth on minimal media without histidine (left). As a toxicity control, yeast harboring these plasmids can grow in media supplemented with histidine (right).
In cellular studies, overexpression of the GTP binding–deficient mutant Rab32T39N caused the condensation of mitochondria at the microtubule organizing centers, as assessed by immunofluorescence techniques (Fig. 8 , A–C). In contrast, overexpression of the wild-type protein had little or no effect on mitochondrial distribution (Fig. 8, D and E). The frequency of mitochondrial collapse was measured in transfected cells (50 cells each) from three separate experiments (Fig. 8 G). Mitochondrial collapse was detected in 56 ± 4% of cells expressing the Rab32T39N mutant (Fig. 8 G), and only 8.6 ± 2% of the wild-type cells exhibited this phenomena (Fig. 8, D, E, and G). Mitochondrial collapse did not appear to involve PKA anchoring because expression of the PKA binding–deficient mutant, Rab32L188P, did not alter the incidence of mitochondrial collapse (Fig. 8 G). In addition, expression of a Rab32T39N, L188P double mutant caused the same degree of mitochondrial collapse as the Rab32T39N mutant (Fig. 8 G). However, correct targeting to the mitochondria appears to contribute to this phenomena, as expression of Rab32T39NΔCC, a GTP binding deficient–mutant lacking the COOH-terminal cysteine pair, displayed a reduced incidence of mitochondrial collapse (32 ± 2%; Fig. 8 G). Because endogenous Rab32 was detected in Cos7 cells (Fig. 6, A and E), our cellular experiments suggest that Rab32T39N acts as a dominant negative mutant. In specificity control experiments, we did not observe any abnormal clustering of endosomes (Fig. 8, H–J), lysosomes (Fig. 8, K–M), or endoplasmic reticulum (Fig. 8, N–P) in cells exhibiting a Rab32T39N-induced mitochondrial collapse.
Figure 8.

The GTP binding mutant Rab32T39N induces mitochondrial collapse around the microtubule organizing center. Cos7 cells were transiently transfected with Flag–Rab32 or Flag–Rab32T39N. Cells were labeled with MitoTracker RedTM for 30 min, fixed, and permeabilized before the Rab proteins were detected with a FITC-conjugated antiFlag antibody. (A) Rab32T39N mutant or (D) Rab32 and (B and E) mitochondria are shown. (C and F) Composite images are presented. A mitochondrial collapse phenotype is observed in Rab32T39N-expressing cells (B, inset). (G) Mitochondrial collapse was scored in cells expressing various Rab32 mutants (indicated below each column). Data are presented as the number of cells exhibiting the phenotype per number of cells transfected. 50 Rab-expressing cells were analyzed in three independent experiments for each construct. Control experiments were performed to determine if expression of Rab32T39N (H, K, and N) alters the morphology or distribution of other organelles. (I, L, and O) Mitochondria were visualized with MitoTracker RedTM. (J) Endosomes were stained with antibodies against the marker protein EEA-1. (M) Lysosomes were stained with antibodies against the marker protein LAMP-1. (P) Endoplasmic reticulums were stained with antibodies against the marker protein calnexin.
Rab proteins can function in membrane fusion, membrane fission, and regulate organelle motility. Our data thus far do not distinguish between these events. Expression of Rab32T39N at low levels promotes an intermediate phenotype in which the mitochondria are elongated and highly interconnected (Fig. 9 , A–D). To further explore this effect, Rab32T39N-transfected cells were subjected to nocodazole treatment for 1 h before fixation (Fig. 9 E). Depolymerization of the microtubules did not result in a dispersion of the mitochondria (Fig. 9, F and G), but there was a preponderance of highly interconnected and elongated mitochondria (Fig. 9 G). Often there were fewer mitochondria per cell, suggesting that expression of Rab32T39N promoted abnormal fusion (unpublished data). In contrast, Cos7 cells expressing wild-type Rab32 are individually dispersed throughout the cytoplasm upon nocodazole treatment (Fig. 9, I–L).
Figure 9.
Expression of Rab32T39N leads to aberrant mitochondrial fusion. Analysis of mitochondrial morphology was performed on cells expressing various levels of Rab32T39N (A–H) and the wild-type protein (I–L). (A) Analysis of collapsed mitochondrial morphology in Cos7 cells expressing low levels of Flag–Rab32T39N. (B) Mitochondria were detected with MitoTracker RedTM stain. (C) A 3× enlargement of the boxed region shown in B (yellow box). (D) Composite image of A and B is presented. (E) Analysis of collapsed mitochondrial morphology in Cos7 cells expressing Flag–Rab32T39N treated with 5 μM nocodazole for 1 h. (F) Mitochondria were detected with MitoTracker RedTM stain. (G) A 3× enlargement of the boxed region shown in F (yellow box). (H) Composite image of E and F is presented. (I) Analysis of dispersed mitochondrial morphology in Cos7 cells expressing Flag–Rab32 treated with 5 μM nocodazole for 1 h. (J) Mitochondria were detected with MitoTracker RedTM stain. (K) A 3× enlargement of the boxed region shown in J (yellow box). (L) Composite image of I and J is presented.
Discussion
Rab32 is a member of the Ras superfamily of small molecular weight G-proteins and belongs to a subclass of these proteins that have a nonconsensus catalytic domain (Bao et al., 2002). We now show that Rab32 functions as an AKAP as it associates with the cAMP-dependent protein kinase (PKA) inside cells. Although we have previously shown that two Rho family GTPases interact with AKAPs (Westphal et al., 2000; Diviani et al., 2001), we provide the first evidence for a direct interaction between a small molecular weight G-protein and the PKA holoenzyme. Interestingly, two other Rab family members interact with protein kinases. Rab8 binds to a GC kinase in a GTP-dependent manner (Ren et al., 1996), and Rab3d copurifies with a ser/thr kinase in the RBL-2H3 mast cell line (Pombo et al., 2001). Our study further emphasizes two additional properties of Rab32: it is localized to mitochondria and it participates in the control of mitochondrial dynamics.
AKAPs are a growing family of functionally related proteins that are classified on the basis of their interaction with the R subunits of the PKA holoenzyme. Common properties include a conserved helical region of 14–18 residues that binds to the R subunit dimer (Carr et al., 1991; Newlon et al., 2001). Our mutagenesis and peptide-binding experiments have located PKA anchoring determinants between residues 178 and 197 of Rab32, which includes the α5 helical region that is conserved in all Ras family members (Bourne et al., 1991). Our modeling studies used the coordinates from the crystal structure of Rab3a to predict that the α5 helix of Rab32 is exposed and available to participate in PKA anchoring. Interestingly, other members of the Ras superfamily utilize this helix in a variety of protein–protein interactions (Maesaki et al., 1999; Morreale et al., 2000). However, only Rab32 and Rab RP-1, a closely related Drosophila orthologue, bind to RII experimentally. This indicates that determinants reside within the α5 helices of both isoforms that accommodate PKA anchoring. Surprisingly, the ability to bind PKA may be determined by a single amino acid side chain. Rab32 and RabRP-1 contain an alanine at position 185 within the helix (numbering based on the Rab32 sequence), whereas most other Rab proteins have a phenylalanine at this position. In fact, substitution with phenylalanine at this site abolishes PKA binding to Rab32, suggesting that a bulky hydrophobic side chain at this site perturbs PKA anchoring. Likewise, introduction of phenylalanine at the corresponding sites in AKAP79, mAKAP, AKAP-KL, AKAP18, or AKAP-Lbc abolishes interaction with RII, as assessed by peptide array analysis (unpublished data). These findings emphasize the high degree of specificity that is built into the PKA anchoring regions.
A proposed function for AKAPs is to direct PKA and other signaling enzymes close to their substrates (Colledge and Scott, 1999; Smith and Scott, 2002). This is generally achieved by targeting domains that direct the AKAP signaling complex to discrete cellular environments. Our subcellular fractionation and immunolocalization data indicate that Rab32 is localized at the mitochondria. Although this is the first report of a Rab protein associated with this organelle, other AKAPs, such as sAKAP84/D-AKAP-1 and D-AKAP-2, are targeted to the outer mitochondrial membranes (Huang et al., 1997a,b). Mitochondrial targeting of sAKAP84/D-AKAP-1 is mediated by a consensus mitochondrial targeting sequence (Lin et al., 1995; Huang et al., 1999). Rab32 does not contain this sequence, but rather has a pair of cysteines at the COOH terminus that are necessary for correct targeting. Both residues are conserved in most Rab proteins and are likely to be sites of lipid modification. Such modifications may be required for insertion and retention of Rab32 in the mitochondrial membrane (Glomset and Farnsworth, 1994). However, additional protein–protein interactions are likely to specify mitochondrial targeting, because high-level expression of Rab32 leads to the detection of the protein throughout the cytoplasm (Fig. 8 D). Overexpression of Rab32 may saturate its “receptor protein” on the mitochondrial membrane, leading to an excess accumulation in the cytoplasm. It is likely that the COOH-terminal hypervariable domain of Rab32 is responsible for such interactions (Chavrier et al., 1991), however the exact mechanism of targeting remains to be determined.
Most Rab proteins cycle between an on state, where GTP is bound, and an off state, where GDP is bound. Accordingly, Rabs can elicit GTP-dependent biological responses by activating a downstream effector. We show that PKA can associate with Rab32 in the presence of GTP or GDP. This is consistent with the notion that Rab32 functions as a conventional AKAP, as there are several examples of anchoring proteins that bind PKA and contain functionally independent domains that possess other biological activities (Westphal et al., 2000; Diviani et al., 2001). Alternatively, there may be multiple pools of Rab32, some of which harbor anchored PKA. This is supported by our immunolocalization data indicating that not all of the mitochondrial-targeted Rab32 is associated with PKA. Thus, the Rab32–PKA complex may participate in distinct cellular processes from the free Rab protein. Defining the molecular interactions regulated by Rab32 should give insight into the PKA function of this complex. For example, specific Rab32 effector proteins, GTPase activating proteins, and GEFs may serve as PKA substrates.
Rab family members participate in membrane trafficking events that are often associated with organelles such as the endoplasmic reticulum, the Golgi network, endosomes, and membrane vesicles. Functions for some of these proteins have been described by genetic analyses in yeast, flies, and mice or by the expression of interfering mutants in mammalian cells. Using the latter approach, we have established that transient transfection of a GTP binding–deficient form, Rab32T39N, selectively induces the collapse of mitochondria at microtubule organizing centers, yet has no effect on the distribution of other organelles (Fig. 8). We propose that the Rab32T39N mutant acts as a dominant negative inhibitor of the endogenous Rab32 protein. Consistent with this notion, endogenous Rab32 is found in Cos7 cells and localizes to the mitochondria (Fig. 6 A). Furthermore, expression of the nonmitochondrial-targeted double mutant Rab32T39NΔCC exhibited a significant reduction in the incidence of mitochondrial collapse. These data suggest that mitochondrial localization of the Rab32T39N mutant contributes to the phenotype. It is possible that Rab32T39N interferes with an upstream activator, such as a GEF, that is localized at mitochondria. This proposition is analogous to the mechanism by which GTP binding–deficient Ras mutants act as dominant inhibitors of specific Ras-GEF proteins (Farnsworth and Feig, 1991).
Our analysis of the Rab32T39N mutant points toward a role for Rab32 in the regulation of mitochondrial fission events, as we detect perinuclear clusters of mitochondria upon overexpression of this interfering mutant. Similar effects are observed in cells expressing an interfering mutant of the dynamin-related GTPase Drp1, which functions to sever the outer mitochondrial membrane in yeast, C. elegans, and mammalian cells (Labrousse et al., 1999; Sesaki and Jensen, 1999; Smirnova et al., 2001). Expression of Drp-1–interfering mutants results in a frequency of collapsed mitochondria, and disruption of the microtubule network with nocodazole reveals a fewer number of elongated organelles (Smirnova et al., 2001). These observations are remarkably similar to the changes in mitochondrial morphology that we recorded upon drug treatment in Rab32T39N-expressing cells. Furthermore, low expression of Rab32T39N reveals an intermediate phenotype in which the mitochondria are partially collapsed. We observed a dense network of seemingly fused mitochondria in these cells. A potential mechanism for the action of the Rab32T39N mutant is to shift the balance of mitochondrial fusion and fission events toward increased fusion. Therefore, the cellular process regulated by Rab32 is most likely mitochondrial division. Future studies will be aimed at elucidating the complement of effector proteins involved in the assembly of a Rab32-mediated mitochondrial fission complex, and the role of anchored PKA in the regulation of these events.
Materials and methods
Yeast two-hybrid
The cDNA from the RIIα dimerization/AKAP-binding domain was PCR amplified and subcloned into the EcoRI-BamHI sites in the pLexA yeast expression vector. This gene encodes an NH2-terminal LexA DNA binding domain fused to the first 45 amino acids of RIIα. 500 μg of a human brain MATCHMAKER cDNA library (CLONTECH Laboratories, Inc.) was screened for AKAPs using the yeast two-hybrid assay as described by Hollenberg et al. (1995).
Cloning full-length Rab32
An EST clone for Rab32 was obtained from Genome Systems (GenBank/EMBL/DDBJ accession no. AI281920; Image 1882928). This clone contained the cDNA sequence representing amino acids 19–225 of Rab32 including the termination codon. A synthetic oligonucleotide was designed to contain the 90 base pairs corresponding to the 5′ region of Rab32 cDNA. The partial Rab32 EST was used as a template for PCR amplification with the 90-mer 5′ primer and a reverse primer that corresponds to the 3′ end of the Rab32 coding sequence. The resulting PCR product was subcloned into the bacterial expression vector pGex4T-1. Full-length Rab32 was confirmed by sequencing.
Construction of expression vectors
The full-length Rab32 cDNA was PCR amplified from the pGEX4T template with synthetic oligonucleotide primers and directly subcloned into mammalian and yeast expression vectors. For the yeast expression vector (pLexA) and the mammalian expression vector (Flag-pcDNA3.1), PCR fragments were subcloned into the 5′ EcoRI sites and 3′ BamHI sites. All Rab32 mutants were generated using Quick-ChangeTM mutagenesis (Stratagene) with specific primers. All PCR reactions were performed using Pfu Turbo polymerase (Stratagene). Each expression construct was sequenced to determine correct reading frame and the existence of appropriate mutations.
Bacterial expression and purification of recombinant Rab32
Recombinant Rab32 and mutants were expressed from the bacterial expression vector pGex4T in the BL-21/DE3 strain of Escherichia coli and purified as NH2-terminal GST fusions using glutathione-Sepharose (Amersham Biosciences).
RII interaction assays
RII overlays were performed using murine [32P]RIIα as previously described (Carr and Scott, 1992). For yeast two-hybrid analysis, the cDNA encoding RIIα 1–45 was subcloned into the Gal4 activation domain containing yeast expression vector pACT2. This gene encodes an NH2-terminal Gal4 activation domain fused to RIIα 1–45. Each pLexA-Rab32, -Rab32L188P, -Rab5, -Rab6, or -Rab7 was cotransformed with the pACT2-RIIα 1–45 into the yeast strain L40 as described above.
Autospot peptide array
Peptide arrays were synthesized on cellulose paper using an Auto-Spot robot ASP222 (ABiMED) as previously described (Frank and Overwin, 1996). After synthesis, the NH2 termini were acetylated with 2% acetic acid anhydride in DMF. The peptides were then deprotected by a 1-h treatment with DCM–TFA, 1:1, containing 3% triisopropylsilane and 2% water.
Immunoprecipitations and PKA activity assay
Cells 50–80% confluent were transfected using Lipofectamine Plus reagents (Invitrogen) according to the manufacturer's instruction. 5 μg of plasmid DNA (Flag–Rab32 or Flag–Rab32L188P) was used to transfect HEK-293 cells in 10-cm dishes. Transfections were performed for 24 h, followed by lysis in IP buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). Recombinant proteins were immunoprecipitated using Sepharose-conjugated antiFlag monoclonal antibody (Sigma-Aldrich). PKA kinase assays were performed as previously described (Corbin and Reimann, 1974).
Northern blot
A Northern blot containing immobilized samples of mRNA from several human tissues (CLONTECH Laboratories, Inc.) was assayed using a 32P-radiolabeled 113-bp probe corresponding to the COOH-terminal hypervariable domain of Rab32. Human RNA dot blot analysis was performed according to the manufacturer's instructions (CLONTECH Laboratories, Inc.).
Western blot analysis and subcellular fractionation of WI-38 cells
Antibodies to Rab32 were raised in rabbits (Covance) against the unique peptide CEENDVDKIKLDQETLRAEN (Princeton Biomolecules). Anti-Rab32 antibodies were affinity purified against recombinant Rab32 coupled to Affigel-10/15 according to the manufacturer's instructions (Bio-Rad Laboratories). WI-38 cell extracts were prepared by lysing confluent cells in buffer (20 mM Hepes, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail [Roche]). Lysates were incubated on ice for 10 min and centrifuged for 20 min at 20,000 g. 20 μg of supernatant was subjected to Western blot analysis. Subcellular fractionation studies were performed as previously described (Lutsenko and Cooper, 1998). In brief, a 10-cm dish of confluent WI38 cells was harvested by trypsin digestion, washed, and resuspended in 1 ml of HB buffer (10 mM Hepes-NaOH, 250 mM sucrose, and protease cocktail, pH 7.5). Cells were dounce homogenized (10–15 strokes) and subjected to differential centrifugation as previously described. 20 μg of proteins from whole cell extract and P1 (nuclear pellet) and P2 (mitochondria enriched) fractions were subjected to Western blot analysis. Control experiments were performed using anti–COX subunit I antibody (Molecular Probes).
Confocal microscopy
Cells were seeded on glass coverslips and incubated overnight at 37°C under 5% CO2. To detect mitochondria, coverslips were incubated with MitoTracker RedTM (Molecular Probes) for 30 min at 37°C under 5% CO2. In recombinant Rab32 localization studies, mitochondria were detected by coexpression of the plasmid pEYFP-Mito (CLONTECH Laboratories, Inc.). RII monoclonal antibodies (Transduction Labs) were used in colocalization studies. Coverslips were washed twice with PBS and fixed for 10 min in PBS–3.7% formaldehyde. Immunocytochemistry was performed as described by Westphal et al. (2000). FITC-, Texas red–, or Cy5-conjugated secondary antibodies were used for detection of the primary antibody (Jackson ImmunoResearch Laboratories). Cells were washed and mounted using the Prolong system (Molecular Probes). Immunofluorescent staining, MitoTracker RedTM, or intrinsic YFP fluorescence was visualized on a Bio-Rad Laboratories MRC1024 UV laser-scanning confocal microscope.
GDP and GTP binding assays
GDP off and GTP on rates were measured as previously described (Self and Hall, 1995). In brief, purified recombinant GST–Rab32 was resuspended in assay buffer (50 mM Tris-HCl, 50 mM NaCl, 5 mM MgCl2, 5 mM DTT, pH 7.5) supplemented with 10 mM EDTA. GDP off rate was determined in high magnesium conditions. GTP on rates were determined in low magnesium conditions. Samples were dissolved in scintillation fluid, followed by counting the radioactive emissions. Data are expressed as the percent nucleotide bound, normalized to time 0 (100%). The nucleotide-dependent RII binding studies were conducted as follows: 1 mg of GST–Rab32 bound to glutathione-Sepharose was nucleotide depleted in 50 mM Tris-HCl, 50 mM NaCl, 10 mM EDTA, 5 mM MgCl2, 5 mM DTT, pH 7.5, followed by loading with 1 mM GDP or 1 mM GTPγS for 30 min at room temperature. The nucleotides were stabilized on Rab32 by extensive washing in high magnesium buffer (50 mM Tris-HCl, 50 mM NaCl, 5 mM MgCl2, 5 mM DTT, pH 7.5, plus either 1 mM GDP or 1 mM GTPγS). 1 μg of recombinant RIIα was incubated with 1 μg nucleotide-loaded Rab32 for 1 h at room temperature, followed by extensive washing in high magnesium buffer. The extent of RII binding was determined by Western blot analysis using a monoclonal anti-RIIα antibody (Transduction Labs).
Rab32 functional experiments
Cos7 cells at 50–80% confluence were transfected with 1 μg of the mammalian expression vectors Flag–Rab32, –Rab32T39N, –Rab32ΔCC, –Rab32T39NΔCC, or –Rab32L188P using Lipofectamine Plus (Invitrogen). Transfected cells were incubated at 37°C, 5% CO2 for 8 h. Cells were labeled with MitoTracker RedTM (Molecular Probes) for 30 min, and processed for immunocytochemistry. Recombinant Rab32 and mutants were detected using a FITC-conjugated antiFlag antibody. 50 Rab32- and mutant-expressing cells were scored for a mitochondrial collapse phenotype, in which the mitochondrial stain was predominantly found at the microtubule organizing center or around the nucleus. Three independent experiments were performed and the results are shown as the percentage of transfected cells with the observed phenotype for each Rab32 construct. To detect cellular markers, primary antibodies against EEA-1 (Transduction Labs), LAMP-1 (Transduction Labs), Calnexin (StressGen Biotechnologies), and GM130 (Transduction Labs) were used in immunocytochemistry assays, and detected by Cy5-conjugated secondary antibodies. Immunofluorescent staining and MitoTracker RedTM was visualized on a Bio-Rad Laboratories MRC1024 UV laser-scanning confocal microscope. For microtubule depolymerization studies, transfected Cos7 cells were treated with 5 μM nocodazole (Sigma-Aldrich) for 1 h before fixation.
Acknowledgments
We thank Dr. Bruno Goud (Institut Curie, Paris, France) for providing the Rab yeast expression vectors and Dr. Philip Stork (Vollum Institute) for providing the Flag-pcDNA3.1 mammalian expression vector. Special thanks go to Lorene Langeberg for confocal microscopy assistance and preparation of the manuscript, Robert Mouton for tissue culture support, and all members of the Scott Lab for insightful discussion and critical review of the manuscript.
This research was supported by DK54441 to J.D. Scott.
Footnotes
Abbreviations used in this paper: AKAP, A-kinase anchoring protein; GEF, guanine nucleotide exchange factor; PKA, cAMP-dependent protein kinase; RIIa, type II regulatory subunit.
References
- Bao, X., A.E. Faris, E.K. Jang, and R.J. Haslam. 2002. Molecular cloning, bacterial expression and properties of Rab31 and Rab32. Eur. J. Biochem. 269:259–271. [DOI] [PubMed] [Google Scholar]
- Bock, J.B., H.T. Matern, A.A. Peden, and R.H. Scheller. 2001. A genomic perspective on membrane compartment organization. Nature. 409:839–841. [DOI] [PubMed] [Google Scholar]
- Bourne, H.R., D.A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 349:117–121. [DOI] [PubMed] [Google Scholar]
- Carr, D.W., and J.D. Scott. 1992. Blotting and band-shifting: techniques for studying protein-protein interactions. Trends Biochem. Sci. 17:246–249. [DOI] [PubMed] [Google Scholar]
- Carr, D.W., R.E. Stofko-Hahn, I.D.C. Fraser, S.M. Bishop, T.S. Acott, R.G. Brennan, and J.D. Scott. 1991. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266:14188–14192. [PubMed] [Google Scholar]
- Chavrier, P., J.P. Gorvel, E. Stelzer, K. Simons, J. Gruenberg, and M. Zerial. 1991. Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature. 353:769–772. [DOI] [PubMed] [Google Scholar]
- Colledge, M., and J.D. Scott. 1999. AKAPs: from structure to function. Trends Cell Biol. 9:216–221. [DOI] [PubMed] [Google Scholar]
- Corbin, J.D., and E.M. Reimann. 1974. A filter assay for determining protein kinase activity. Methods Enzymol. 38:287–294. [DOI] [PubMed] [Google Scholar]
- Diviani, D., J. Soderling, and J.D. Scott. 2001. AKAP-Lbc anchors protein kinase A and nucleates Gα 12-selective Rho-mediated stress fiber formation. J. Biol. Chem. 276:44247–44257. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam, B., S. Guo, and K.J. Kemphues. 1995. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 83:743–752. [DOI] [PubMed] [Google Scholar]
- Farnsworth, C.L., and L.A. Feig. 1991. Dominant inhibitory mutations in the Mg(2+)-binding site of RasH prevent its activation by GTP. Mol. Cell. Biol. 11:4822–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciello, A., M.E. Gottesman, and E.V. Avvedimento. 2001. The biological functions of A-kinase anchor proteins. J. Mol. Biol. 308:99–114. [DOI] [PubMed] [Google Scholar]
- Frank, R., and H. Overwin. 1996. SPOT synthesis. Epitone analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol. Biol. 66:149–169. [DOI] [PubMed] [Google Scholar]
- Glomset, J.A., and C.C. Farnsworth. 1994. Role of protein modification reactions in programming interactions between ras-related GTPases and cell membranes. Annu. Rev. Cell Biol. 10:181–205. [DOI] [PubMed] [Google Scholar]
- Hausken, Z.E., V.M. Coghlan, C.A.S. Hasting, E.M. Reimann, and J.D. Scott. 1994. Type II regulatory subunit (RII) of the cAMP dependent protein kinase interaction with A-kinase anchor proteins requires isoleucines 3 and 5. J. Biol. Chem. 269:24245–24251. [PubMed] [Google Scholar]
- Hollenberg, S.M., R. Sternglanz, P.F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L.J., K. Durick, J.A. Weiner, J. Chun, and S.S. Taylor. 1997. a. D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc. Natl. Acad. Sci. USA. 94:11184–11189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L.J., K. Durick, J.A. Weiner, J. Chun, and S.S. Taylor. 1997. b. Identification of a novel dual specificity protein kinase A anchoring protein, D-AKAP1. J. Biol. Chem. 272:8057–8064. [DOI] [PubMed] [Google Scholar]
- Huang, L.J., L. Wang, Y. Ma, K. Durick, G. Perkins, T.J. Deerinck, M.H. Ellisman, and S.S. Taylor. 1999. NH2-terminal targeting motifs direct dual specificity A-kinase–anchoring protein 1 (D-AKAP1) to either mitochondria or endoplasmic reticulum. J. Cell Biol. 145:951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, T. 2000. Signaling--2000 and beyond. Cell. 100:113–127. [DOI] [PubMed] [Google Scholar]
- Joberty, G., C. Petersen, L. Gao, and I.G. Macara. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531–539. [DOI] [PubMed] [Google Scholar]
- Jordan, J.D., E.M. Landau, and R. Iyengar. 2000. Signaling networks: the origins of cellular multitasking. Cell. 103:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchinke, U., F. Grawe, and E. Knust. 1998. Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr. Biol. 8:1357–1365. [DOI] [PubMed] [Google Scholar]
- Labrousse, A.M., M.D. Zappaterra, D.A. Rube, and A.M. van der Bliek. 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 4:815–826. [DOI] [PubMed] [Google Scholar]
- Lin, D., A.S. Edwards, J.P. Fawcett, G. Mbamalu, J.D. Scott, and T. Pawson. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547. [DOI] [PubMed] [Google Scholar]
- Lin, R.-Y., S.B. Moss, and C.S. Rubin. 1995. Characterization of S-AKAP84, a novel developmentally regulated A kinase anchor protein of male germ cells. J. Biol. Chem. 270:27804–27811. [DOI] [PubMed] [Google Scholar]
- Lutsenko, S., and M.J. Cooper. 1998. Localization of the Wilson's disease protein product to mitochondria. Proc. Natl. Acad. Sci. USA. 95:6004–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maesaki, R., K. Ihara, T. Shimizu, S. Kuroda, K. Kaibuchi, and T. Hakoshima. 1999. The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1. Mol. Cell. 4:793–803. [DOI] [PubMed] [Google Scholar]
- Morreale, A., M. Venkatesan, H.R. Mott, D. Owen, D. Nietlispach, P.N. Lowe, and E.D. Laue. 2000. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 7:384–388. [DOI] [PubMed] [Google Scholar]
- Newlon, M.G., M. Roy, D. Morikis, Z.E. Hausken, V. Coghlan, J.D. Scott, and P.A. Jennings. 1999. The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat. Struct. Biol. 6:222–227. [DOI] [PubMed] [Google Scholar]
- Newlon, M.G., M. Roy, D. Morikis, D.W. Carr, R. Westphal, J.D. Scott, and P.A. Jennings. 2001. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 20:1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick, P., and M. Zerial. 1997. The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol. 9:496–504. [DOI] [PubMed] [Google Scholar]
- Ostermeier, C., and A.T. Brunger. 1999. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 96:363–374. [DOI] [PubMed] [Google Scholar]
- Pawson, T. 1995. Protein modules and signalling networks. Nature. 373:573–580. [DOI] [PubMed] [Google Scholar]
- Pombo, I., S. Martin-Verdeaux, B. Iannascoli, J. Le Mao, L. Deriano, J. Rivera, and U. Blank. 2001. IgE receptor type I-dependent regulation of a Rab3D-associated kinase: a possible link in the calcium-dependent assembly of SNARE complexes. J. Biol. Chem. 276:42893–42900. [DOI] [PubMed] [Google Scholar]
- Qiu, R.G., A. Abo, and G.S. Martin. 2000. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCzeta signaling and cell transformation. Curr. Biol. 10:697–707. [DOI] [PubMed] [Google Scholar]
- Ren, M., J. Zeng, C. De Lemos-Chiarandini, M. Rosenfeld, M. Adesnik, and D.D. Sabatini. 1996. In its active form, the GTP-binding protein rab8 interacts with a stress-activated protein kinase. Proc. Natl. Acad. Sci. USA. 93:5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self, A.J., and A. Hall. 1995. Measurement of intrinsic nucleotide exchange and GTP hydrolysis rates. Methods Enzymol. 256:67–76. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and R.E. Jensen. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., L. Griparic, D.L. Shurland, and A.M. van der Bliek. 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 12:2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, F.D., and J.D. Scott. 2002. Signaling complexes: junctions on the intracellular information super highway. Curr. Biol. 12:R32–R40. [DOI] [PubMed] [Google Scholar]
- Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153–208. [DOI] [PubMed] [Google Scholar]
- Westphal, R.S., S.H. Soderling, N.M. Alto, L.K. Langeberg, and J.D. Scott. 2000. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 19:4589–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]