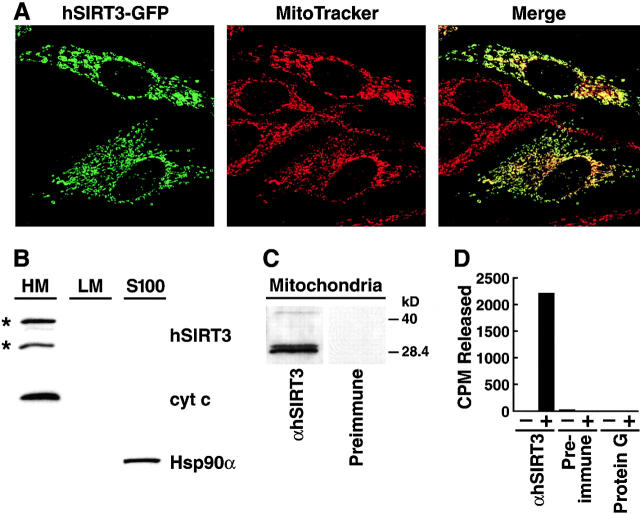
Figure 2.
Subcellular localization of hSIRT3 in mitochondria. (A) hSIRT3–GFP was transfected into HeLa cells grown on coverslips. Cells were stained with the mitochondrial marker MitoTracker, embedded, and analyzed by confocal laser scanning microscopy. (Left) Fluorescence from the hSIRT3–GFP fusion protein. (Middle) Fluorescence from the MitoTracker–stained mitochondria in the same focal plane. (Right) Merged image showing complete overlap of the two staining patterns. (B) HEK293T cells transfected with hSIRT3–FLAG were homogenized and fractionated by differential centrifugation. Equal amounts (30 μg) of heavy membranes (HM), light membranes (LM) and cytosolic proteins (S-100) fraction were analyzed by immunoblotting. hSIRT3–FLAG was revealed by detection with monoclonal M2 anti-FLAG antibodies. Two hSIRT3–FLAG specific forms (asterisks) were detected. Nitrocellulose membranes were stripped and reprobed with antibodies against cytochrome c (cyt c) and Hsp90α. (C) Mitochondria were prepared from HEK293T cells and lysates were analyzed by Western blotting with a polyclonal rabbit hSIRT3 antiserum or preimmune serum obtained from the same rabbit. (D) hSIRT3 was immunoprecipitated from HEK293T cells with hSIRT3 antiserum (0.35 mg/ml), preimmune serum (0.35 mg/ml), or protein G Sepharose. Equal amounts of immunoprecipitate were analyzed for in vitro deacetylase activity in the absence (−) or presence (+) of NAD.