Figure 6.
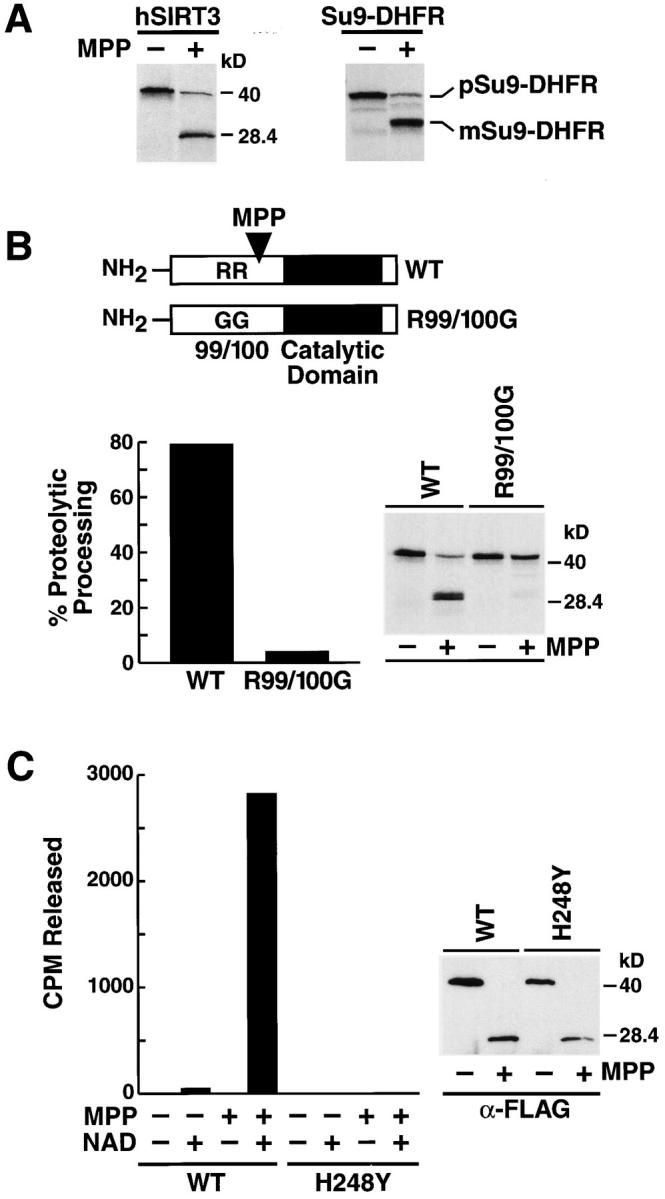
Proteolytic processing of hSIRT3 by MPP leads to enzymatic activation. (A) [35S]-labeled hSIRT3–FLAG (left) or pSu9–DHFR (right) was incubated with purified recombinant yeast MPP for 45 min at 27°C. Samples were analyzed by SDS-PAGE and autoradiography. m, mature form of pSu9–DHFR; p, precursor form. (B) [35S]-labeled hSIRT3–FLAG (WT) and hSIRT3R99/100G–FLAG (R99/1006) were incubated with MPP and analyzed as in A. (Left) Efficiency of proteolytic processing by recombinant yeast MPP was quantitated by phosphorimaging. (Right) Autoradiography of the same experiment. (C) Unlabeled hSIRT3–FLAG (WT) or hSIRT3H248Y–FLAG (H2484) synthesized in vitro in rabbit reticulocyte lysates was incubated with recombinant yeast MPP for 45 min at 27°C. FLAG-tagged proteins were immunoprecipitated with anti-FLAG M2-agarose beads and analyzed for deacetylase activity in vitro with the H4 histone peptide assay in the presence or absence of NAD (1 mM; left). Western blot analysis of immunoprecipitates used in the deacetylase assay (right).
