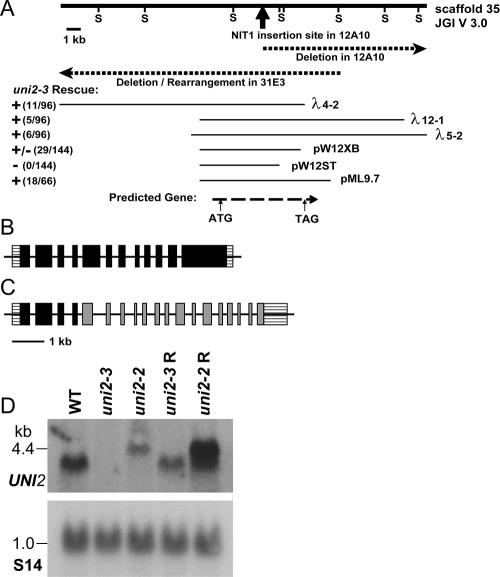
Figure 1.
UNI2 genomic region and transcripts. (A) The heavy solid line depicts a 25-kb genomic segment. Sac1 sites (S) were used in the mapping of genomic DNA from the 12A10 and 31E3 strains. The dark arrow indicates the site of insertion of the pMN24 plasmid DNA in the 12A10 strain. Dashed lines show the genomic region disrupted in the 12A10 and 31E3 strains. Thin lines below show cloned genomic fragments capable (+) and incapable (−) of rescuing the phenotype of the 31E3 strain. The bottom dashed line indicates the location of the coding region of the UNI2 gene as identified by RT-PCR experiments. (B) Schematic showing the structure of the WT UNI2 gene with 12 exons. (C) Schematic showing uni2-2 lesion in which the fourth intron of the UNI2 gene (black) is fused to the second intron of the NIT1 gene (gray). (D) Autoradiographs of RNA blots. Polyadenylated RNA (10 μg/lane) from the indicated cells was electrophoresed on a denaturing gel and transferred to a membrane. (top) The RNA was hybridized with a labeled probe consisting of the second exon of the UNI2 gene. (bottom) A labeled fragment of the RPS14 gene encoding the ribosomal protein S14 was used as a hybridization control for equal loading (Nelson et al., 1994).