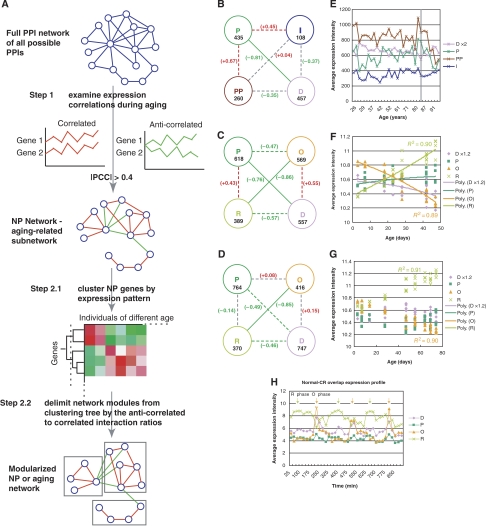
Figure 1.
Network modules during human brain and fruitfly aging. (A) Flow diagram of the NP analysis arriving at the aging-related NP network and modularized NP network. The first step of the NP analysis is to obtain all the PPIs between interactors that are negatively or positively correlated at the transcription level (∣PCC∣>0.4) during aging. Once the subnetwork is found, the genes in the subnetwork are clustered by their expression profiles using hierarchical clustering, then the best separated modules on the clustering tree are found judged by the percentage of negatively correlated interacting gene pairs within a module (<1%). PCC stands for Pearson correlation coefficient. (B) Transcriptional relationships among the modules of the human brain aging NP network. Human D (differentiation), P (proliferation), PP (protein processing) and I (immunity) modules are represented with nodes of lavender, green, brown and dark blue, respectively. Pearson correlation coefficient (PCC) between the two serials of the average expression levels of a pair of the modules over different samples are used to measure the similarity of expression patterns between the modules. The PCC for each module pair is marked at the line connecting the two modules. Solid red and green lines represent strong transcriptional correlations and anticorrelations, respectively (∣PCC∣>=0.7), whereas dotted red and green lines indicate weak correlations and anticorrelations, respectively (0.4<∣PCC∣<0.7). Gray dotted lines represent no obvious transcriptional relationships (∣PCC∣<=0.4). The number of genes of each network module is indicated below the name of the module. (C, D) Transcriptional relationships among the modules of the fruitfly aging under normal (C) and CR (D) conditions. (E) Average expression intensities are plotted against the age of the human subjects for the human brain module genes. The gray vertical line marks age 85. (F, G) Average expression intensities are plotted against fly age for the fly module genes under normal (E) and CR condition (F), respectively. Polynomial fits of the fly module expression level across different populations over age are also displayed, indicating perfect linearity of O and R module expression level with age. Linear regression R2 of the O and R modular changes with age are also displayed. Only the expression levels of the genes overlapping between the corresponding normal diet and CR modules are plotted. Those for the nonoverlapping genes are shown in Supplementary Figure 4. (H) Relationship of O and R modules to metabolic cycle. The average expression levels of the yeast orthologs of the fly R and O genes oscillate with the metabolic cycles and reach the highest levels in the antiphases of reductive and oxidative metabolisms, respectively (marked by the arrows). The P and D modules are also included for comparison. Genes for each module are the overlapping genes between the corresponding modules under normal diet and CR.