Figure 2.
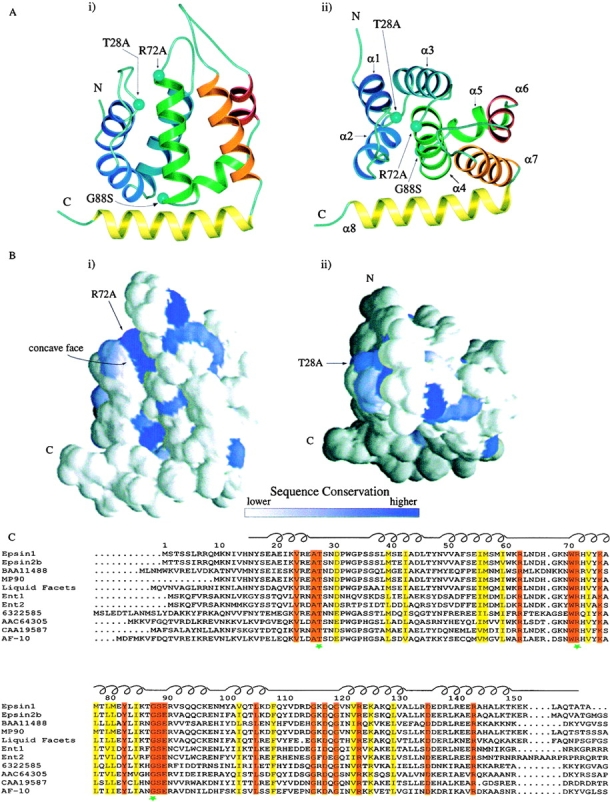
Overall structure of the ENTH domain and sequence alignment. A, Ribbon drawings of the ENTH domain. The drawing at right (ii) is related to the drawing at left (i) by a 60° rotation towards the viewer. The eight α helices of the ENTH domain are indicated by different colors. The T28A, R72A, and G88S mutations made in this study are indicated by aquamarine balls. α Helices 1–8 are indicated in i. Figures generated using MOLSCRIPT (Kraulis 1991). B, Sequence conservation projected onto accessible surface representations of the ENTH domain. These surface representations correspond to the same two views (i and ii) in A. Figures generated using GRASP (Nicholls, 1991). C, Sequence alignment of ENTH domains from various species. Identical residues are indicated in red, conserved residues in yellow. Green stars indicate residues that have been mutated in this study. The schematic diagram above the alignment indicates the location of loop and helix regions. Accession number and species as follows: epsin 1, AAC33823, Rattus norvegicus; epsin 2b, AAC78609, Homo sapiens; BAA11488, Homo sapiens; MP90, AAC60123, Xenopus laevis; liquid facets, AAF05113, Drosophila melanogaster; Ent1, 6320039, S. cerevisiae; Ent2, 6323235, S. cerevisiae; 6322585, S. cerevisiae; AAC64305, Arabidopsis thaliana; CAA19587, Schizosaccharomyces pombe; AF-10, AAB68030, Avena fatua. Alignment generated using CLUSTAL W (Thompson et al. 1994) and AMAS (Livingston and Barton 1993).
