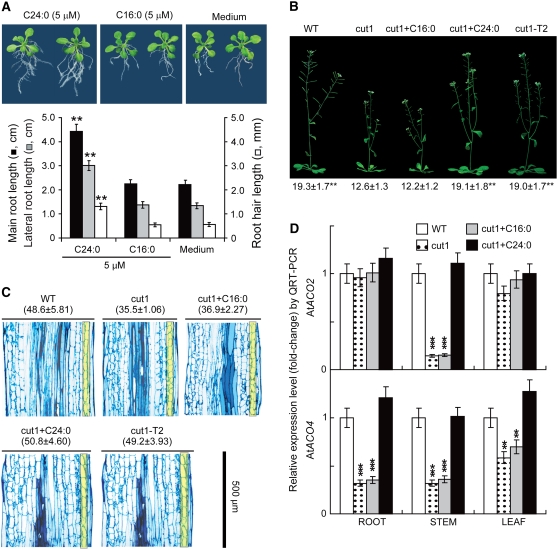
Figure 7.
VLCFA Results in Significant Extension of Several Arabidopsis Cell Types.
(A) Exogenous C24:0 promoted significant Arabidopsis root cell elongation. Top panel, photographs of 14-d-old seedlings grown in medium supplemented with C24:0 or C16:0. Medium, plants received MTBE with no C24:0 or C16:0 added. Bottom panel, statistics obtained from 30 individual seedlings representing three independent experiments. See Figure 1 legend for statistical procedures and annotations. Significance values were obtained by comparison with the Medium group.
(B) Wild-type lengths of stems were produced in Arabidopsis cut1 mutant when C24:0 was applied exogenously through the root system or when Gh KCS13/CER6 was transformed in the mutant background. WT, wild-type Landsberg erecta ecotype plants; cut1+C16:0, cut1 plants that received 50 mL of 5 μM C16:0 supplementation daily from the day of bolting for each 8-inch pot; cut1+C24:0, cut1 plants that received the same amount of 5 μM C24:0; cut1-T2, homozygous transgenic cut1 plants that expressed the Gh KCS13/CER6 gene. The same amount of water containing MTBE was applied to the medium. Photographs were taken of 40-d-old soil-grown plants. Average stem lengths (centimeters) measured from 30 individual plants from three independent experiments are reported beneath representative plants.
(C) Comparisons of longitudinal cell lengths from mature stem sections of wild-type, cut1, cut1+C16, cut1+C24, and cut1-T2 plants. Longitudinal sections were prepared from Arabidopsis stems, and average cell length (micrometers, means ± se; n = 5), reported at the top of each section in parentheses, was calculated by dividing the section length (500 μm) by the number of cells in the cortex layer (shown in yellow). The width of each section roughly reflects the actual thickness of the stem.
(D) C24:0 induced a specific and significant increase in expression of At ACO genes in cut1 root, stem, and leaf cells. The cut1 mutant plants were watered with 50 mL of 5 μM C24:0 or C16:0 for 3 d starting from the day of bolting and were harvested separately as roots, stems, and leaves for RNA extraction. QRT-PCR analyses were performed in triplicate using RNA samples prepared from independent plant materials and gene-specific primers, as listed in Supplemental Table 1 online. Wild-type plants and T2 transgenic plants received the same amount of MTBE (∼0.8 μM). We followed www.ncbi.nlm.nih.gov for nomenclature for At ACOs.