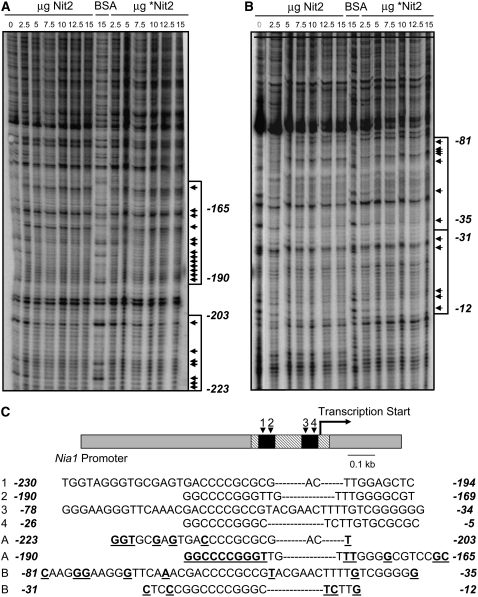
Figure 6.
DNase I Footprinting of Nit2 Protein on the NIA1 Promoter Region.
(A) Patterns of fragments resulting from digestion with DNase I of the 5′ end 32P-labeled DNA fragment upon binding with no protein, with different amounts of full-length Nit2p or Nit2 protein without GAF domain (*Nit2), or with BSA. Sequences protected from DNase I are indicated by arrows (band identification was performed by sequencing; data not shown).
(B) Patterns of fragments resulting from digestion with DNase I of the 3′ end 32P-labeled DNA fragment. The regions of DNA protected from DNase I digestion by Nit2 or truncated *Nit2 proteins are indicated by arrowheads.
(C) NIA1 gene promoter scheme. Oblique hatching shows the DNA fragment used in footprinting assays. Black boxes indicate two regions of the promoter that are essential to full expression of the gene (Loppes and Radoux, 2002). The palindromic sequences proposed as sequence signatures for the DNA binding sites in experiments A and B are compared with those present in the NIA1 gene promoter. Bases showing a clear modified pattern in DNase I digestions are shown in boldface and underlined.