Figure 5.
MIRNA Loops Overaccumulate in fry1 and xrn2 xrn3 Double Mutants.
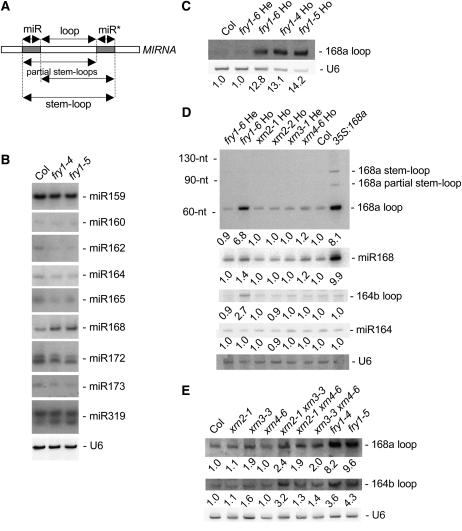
(A) Diagram of the various intermediates and end products generated during MIRNA maturation. The stem-loop is the RNA sequence flanked at each end by the miRNA and miRNA* sequences; partial stem-loops are generated after either the 3′ end of the miRNA or the 5′ end of miRNA* is defined. The positions of the 21-nucleotide miRNA (miR) and miRNA* (miR*) species are indicated by gray boxes (the miR* is the 21-nucleotide RNA species that is opposite of and paired to the miRNA in the MIRNA hairpin). The loop sequence is between the miRNA and the miRNA* and is liberated during miRNA maturation.
(B) RNA gel blot analysis of miRNA accumulation in Col control and fry1-4 and fry1-5 mutant inflorescences. miRNA accumulation is either unchanged or slightly reduced in fry1 mutants except for miR168, which slightly overaccumulates. Blots were stripped and rehybridized with a probe complementary to U6 RNA as a loading control.
(C) RNA gel blot analysis showing that a homozygous fry1-6 T-DNA insertion mutant and fry1-4 and fry1-5 ethyl methanesulfonate mutants overaccumulate MIR168a loops in inflorescence tissues. U6 hybridization is shown as a loading control. Normalized values of the MIR168a loop RNA to U6 snRNA, with Col levels set at 1.0, are indicated.
(D) RNA gel blot analyses of MIR168a and MIR164b maturation intermediates and end products in inflorescence tissues of controls (Col and MIR168a-overexpressing plants; 35S:168a), hemizygous and homozygous fry1 mutants, two homozygous xrn2 mutants, a hemizygous xrn3 mutant, and a homozygous xrn4 mutant. Homozygous fry1 mutants overaccumulate MIR168a and MIR164b loops but not stem-loops or partial stem-loops. miR164 and miR168 accumulation in fry1 homozygous mutants is shown in (B). RNA extracted from MIR168a-overexpressing inflorescences was used to indicate the positions of MIR168a stem-loops and partial stem-loops, which are undetectable in wild-type inflorescences. The positions of 32P-labeled RNA oligonucleotides (nt) are noted. U6 hybridization is shown as a loading control. Normalized values of loop RNAs and miRNAs to U6 snRNA, with Col levels set at 1.0, are indicated.
(E) RNA gel blot analysis of MIR168a and MIR164b loop accumulation in control Col, xrn2, hypomorphic xrn3, xrn4, xrn2 xrn3, xrn2 xrn4, xrn3 xrn4, and fry1 mutant inflorescences. All mutations are homozygous. xrn3-containing mutants overaccumulate loops, although to a lesser extent than fry1 mutants. U6 hybridization is shown as a loading control. Normalized values of loop RNAs to U6 snRNA, with Col levels set at 1.0, are indicated.