Fig. 2.
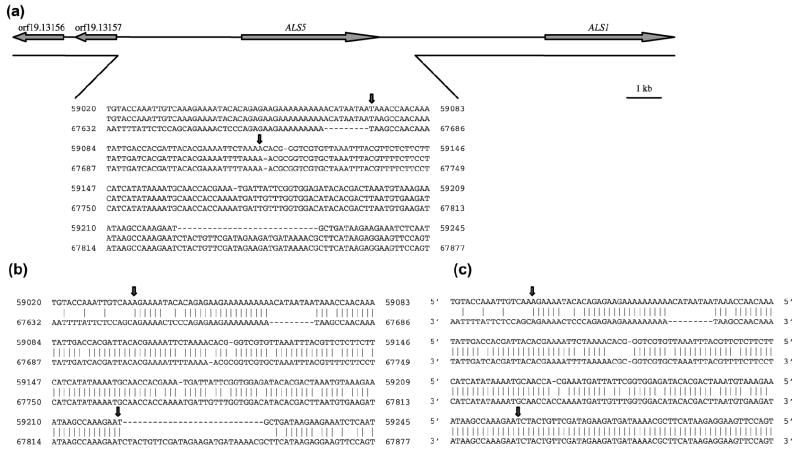
(a) Scale drawing of the chromosome 6 region that encodes ALS5 to indicate the sequences deleted in naturally occurring ALS5-negative strains. DNA sequence information was taken from the Candida Genome Database (http://www.candidagenome.org) Contig 19-20233; sequence coordinates are shown. Deletion of ALS5 is mediated by direct repeats that are 5’ and 3’ of ALS5. The gap in the line below the schematic chromosome indicates the deleted region, which extends from approximately 3.5 kb upstream of ALS5 to 1.0 kb 3’ of the ALS5 stop codon. The total size of the deleted fragment depends upon the size of the ALS5 allele encoded on that chromosome. An ALS5-negative strain has undergone recombination events to remove both alleles from the genome. Alignment of the direct repeat sequence from 5’ of ALS5 (top line; coordinates 59020 to 59425) and 3’ of ALS5 (bottom line; coordinates 67632 to 67877), and the sequence derived from an ALS5-negative clinical isolate (middle line), is shown. Vertical arrows above the sequence alignment delimit the region in which recombination occurs between the 5’ and 3’ direct repeats. (b) Same DNA sequences from upstream and downstream of ALS5 that were shown in (a), but with vertical lines to indicate sequence identity within the region of ALS5 deletion. Vertical arrows delimit the region used to calculate percentage identity (85.3%) between the sequences. Calculation of the length of the direct repeat sequences is complicated because regions of high identity are interspersed with sequence gaps. (c) DNA sequences from the same region in a clade I isolate (see Materials and methods). The similar percentage identity for these sequences (84.7%) to that for the sequences in (b) suggests that recombination to delete ALS5 should be able to occur in this isolate. The higher percentage of sequence identity 3’ of the region delimited by arrows suggests an even higher recombination potential in this strain. It is unclear why strains in clade I have not accumulated deleted ALS5 alleles similar to strains in other clades.