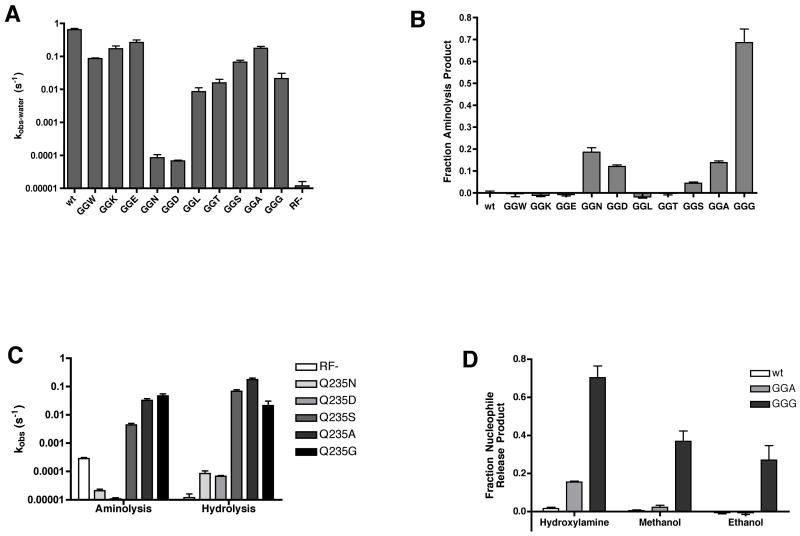
Figure 4. Mutations at Q235 reveal a correlation between side-chain size and water specificity.
(A) Hydrolysis rates of RF1 Q235 mutants presented on a log-scale plot. For mutants where detectable aminolysis product was observed (GGN, GGD, GGS, GGA, and GGG) the hydrolysis rate constant (kobs-water) was calculated from kobs × hydrolysis endpoint. For all other mutants, kobs = kobs-water. Rate constants were measured at least twice and are presented as mean +/− standard error.
(B) Endpoints of aminolysis reactions with a series of RF1 Q235 mutations. Endpoints were calculated as the fraction of aminolysis product to total release product, normalized to the wild type background aminolysis, and are presented as the mean +/− standard error of at least two measurements.
(C) Log-scale plot of rate constants for aminolysis and hydrolysis of RF1 Q235 mutants that exhibit increased aminolysis endpoints. The rate constants for aminolysis and hydrolysis are calculated from their respective endpoints multiplied by kobs. The mean +/− standard error of at least two measurements are presented.
(D) Aminolysis or alcoholysis endpoints of wild type, Q235A and Q235G RF1 measured in the presence of 1 M hydroxylamine, 10% methanol or 10% ethanol. Endpoints are measured as the fraction of aminolysis or alcoholysis product to total release product and presented as the mean +/− standard error of at least three measurements.