Figure 4.
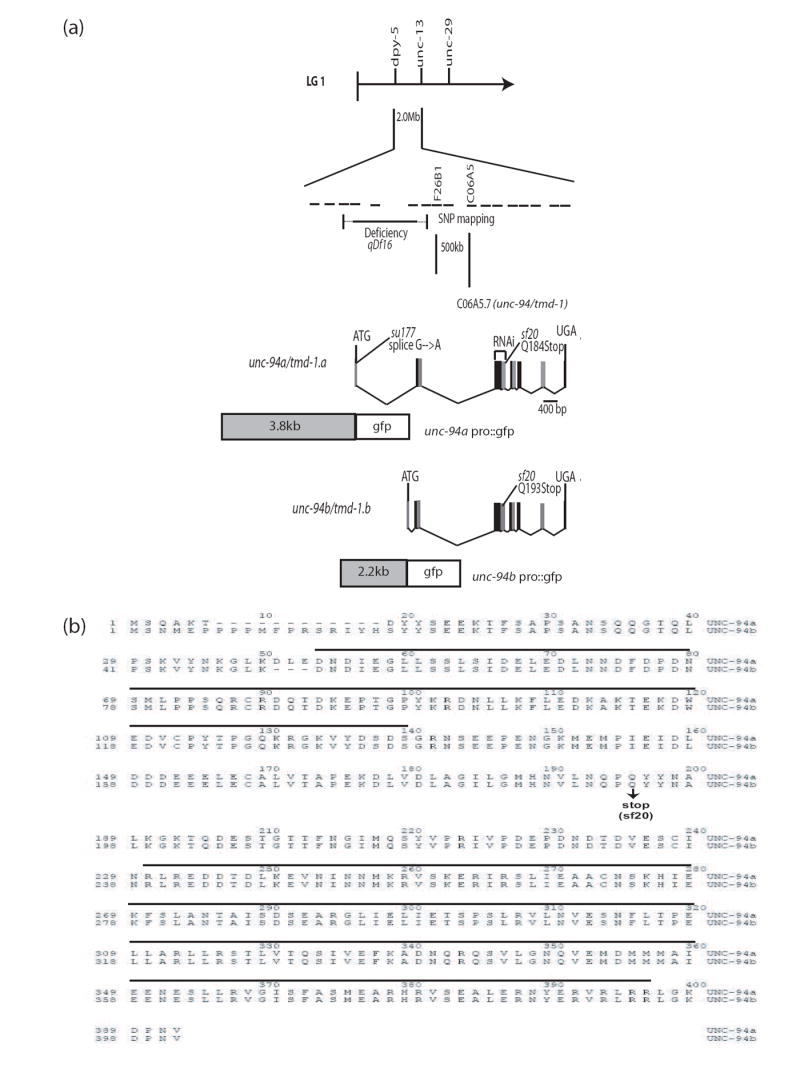
Genetic and physical mapping of unc-94, location of mutation sites for su177 and sf20, and the sequences of UNC-94a and b. (a) By using a combination of deficiency and SNP mapping, unc-94 was placed within a 500 kb region between/within cosmids F26B1 and C06A5. (Each dashed line represents the span of one cosmid insert.) After scanning the region for candidate genes, cosmid C06A5 was injected into unc-94 (su177) animals and was able to rescue the Unc-94 mutant phenotype. When rrf-3 animals were fed dsRNA for C06A5.7, their progeny showed a polarized light phenotype similar to that of unc-94 (su177 and sf20). As shown, WormBase predicts two isoforms for C06A5.7, a and b. In the figure, for clarity consecutive exons are represented as black or grey boxes. After having sequenced both unc-94 alleles, it was noted that su177 contains a splice site mutation in the first intron for isoform a, while sf20 contains a nonsense mutation in the fifth exon, which is shared among the two isoforms. Putative promoter regions plus the first exon of each isoform were used to create promoter-gfp fusions in order to analyze expression patterns. (b) Alignment of UNC-94 a and b protein sequences. The UNC-94 proteins are identical except in two places: (1) at their N-termini because of different first exons, and (2) beginning at residue 39 of UNC-94a which has an additional 3 residues (DLE) not found in UNC-94b because the 5’ end of exon 3 in UNC-94a is 9 bp earlier than in UNC-94b. Indicated is the mutation site of sf20: it is a C-to-T transition which converts glutamine 184 of isoform a (and glutamine 193 of isoform b) to a stop codon (CAA to TAA). The horizontal black lines indicate a putative tropomyosin-binding domain in the amino-terminal half, and a putative actin-binding domain in the carboxy-terminal half of each protein.
