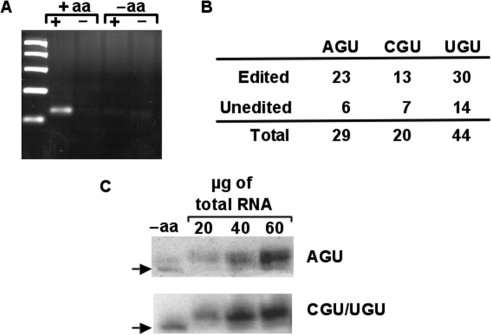
Figure 3.
All tRNAThr isoacceptors are substrate for aminoacylation in vivo. (A) Total RNA from T. brucei was extracted under acidic conditions and aminoacylation levels were correlated to editing levels by the oxopap assay as described. (B) Independent clones derived from the ‘+aa’ reaction (in A) were used in the OXOPAP assay (Materials and Methods section) and analyzed by sequencing. In all cases both edited and unedited species are substrates for aminoacylation in vivo. (C) Total RNA from the same fraction as above was also separated by acid denaturing polyacrylamide electrophoresis and probed with radioactive oligonucleotides specific for either tRNAThrAGU (top panel) or tRNAThrCGU/UGU. The probe used does not discriminate between the CGU and UGU isoacceptors. ‘−aa’ refers to a control reaction in which the RNA was deacylated by incubating under basic conditions prior to analyses. ‘+aa’ refers to the aminoacylated fractions purified and kept under acid pH.