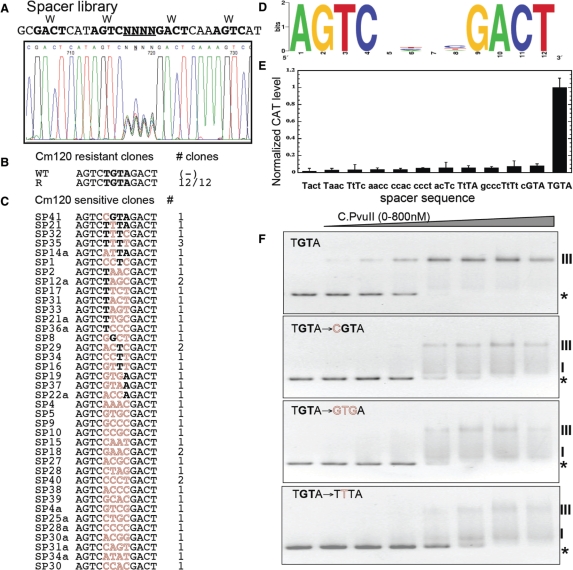
Figure 4.
Randomized operator spacer library and selected variants. (A) Sequencing trace of the pooled randomized spacer plasmid library before selection. The randomized spacer lies between C-half-boxes 1B and 2A. The promoter region library is upstream of a promoterless cat gene (chloramphenicol acetyltransferase). (B) The plasmid library was used to transform an E. coli strain that already carried a compatible plasmid producing C.PvuII at physiological levels, and transformants were plated onto agar with a high Cml concentration (120 µg/ml) to select functional C.PvuII-activated variants, followed by a screen for reduced resistance in the absence of C.PvuII. The sequencing result is shown, along with the WT sequence for comparison. (C) Non-activated variants, that grew at a low Cml concentration (20 µg/ml) but not at 120 µg/ml. Sequences are shown, with matches to the WT sequence in bold, and alterations in outline. (D) Logo analysis of the Cml-sensitive variants shown in (C). (E) Promoter activity for selected variants in the presence of physiological steady-state levels of C.PvuII (pvuIIC under native control from plasmid pDK200). CAT levels were determined in triplicate via immunoassay as described in Material and Methods section. Error bars indicate the SD. CAT levels are expressed as nanograms of protein per microgram of total protein and then normalized to the WT PpvuIICR-cat level (TGTA). (F) EMSA performed for WT (TGTA) and three representative variants. The experiments were carried out as in Figure 3B. The stars and Roman numerals denote positions of unbound DNA and C.PvuII–DNA complexes, respectively.