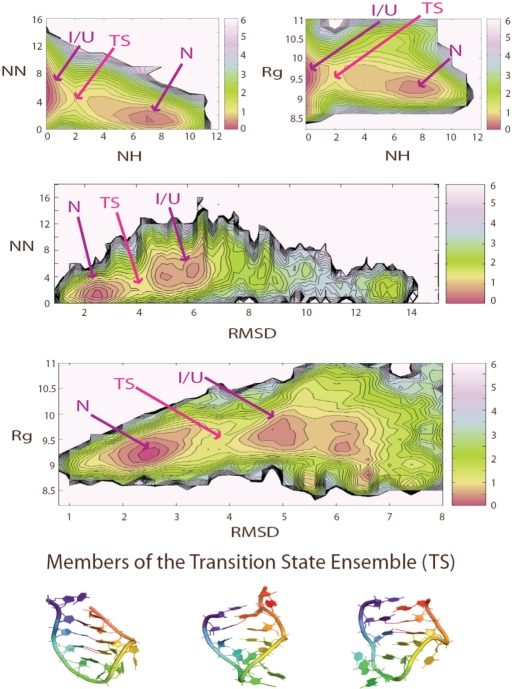
Figure 3.
The PMFs of the RNA hairpin at 300 K are shown as a function of a number of order parameters. NH is the number of native hydrogen bonds and NN the number of non-native hydrogen bonds. Rg is the radius of gyration of the hairpin and RMSD is the root mean square deviation from the hairpin crystal conformation in the ribosome. The crystal structure has 12 native hydrogen bonds: 3 belong to the U-turn sub-motif, 2 belong to the U-A trans WC/H bp and 7 belong to the helical stem. Local minima are indicated in red. The transition state is defined as the barrier region between the native and non-native basins. The energy of the native and non-native basin is similar, and the energy of the transition state is at least 1.5 kcal/mol higher than the two basins. Sample transition state structures, which contain 1–3 native hydrogen bonds in the helical stem, are shown here. All RMSD and Rg values are in Angstroms units (Å). PMF is plotted in units of kcal/mol, where 1 kcal roughly equals to 0.6 KT at 300 K.