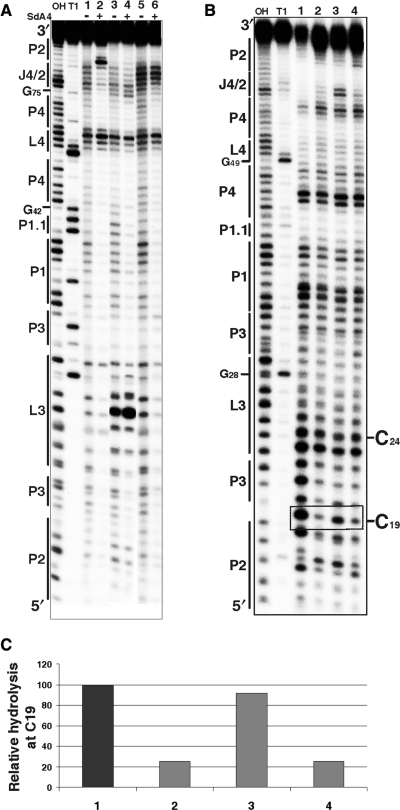
Figure 4.
Chemical and enzymatic probing of the bottom of the P2 stem. (A) Autoradiogram of a 10% PAGE gel of in-line probing performed on 5′-end-labelled wild-type and mutated trans-acting ribozymes. Both alkaline and RNase T1 hydrolyses of the wild-type ribozyme were performed in order to determine the location of each position (lanes OH and T1, respectively). In-line probing of the wild-type ribozyme (1, 2), the RzC24U,C25U,G40U,G41U (3, 4) and the RzA78U,A79U (5, 6) mutants are shown. The experiments were performed either in the absence (−) or the presence (+) of the SdA4 analogue. The secondary structure motifs are identified on the left. (B) Autoradiogram of a 10% PAGE gel of RNase V1 probing performed on 5′-end-labelled wild-type and mutated cis-acting ribozymes. Both alkaline and RNase T1 hydrolyses of the wild-type sequence were performed in order to determine the location of each position (lanes OH and T1, respectively). Lanes 1 to 4 correspond to the wild-type sequence (C19G81G80) and the mutants RzC19,G81A,G80A, RzC19,G81AG80 and RzC19G81G80A, respectively. The positions of the C19 and C24 (used to establish the relative level of hydrolysis) are indicated on the right. (C) Histogram of the relative levels of RNase V1 hydrolysis of C19 for each ribozyme.