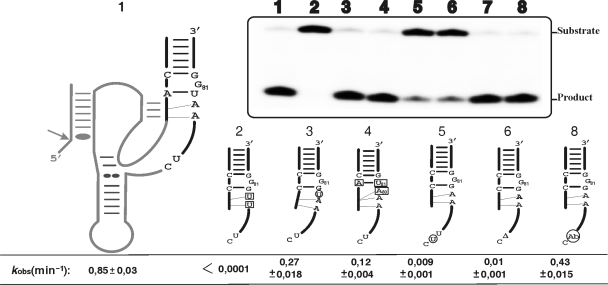
Figure 6.
Cleavage activity assays of various mutants of the J4/2 junction. The inset shows a typical autoradiogram of a PAGE gel for a 30 min reaction for each ribozyme tested. The secondary structure of the wild-type ribozyme is illustrated entirely on the left, while only the P2 stem, J4/2 junction and the most relevant nucleotides are illustrated for the mutants. The mutations and insertion are denoted by squared and circled nucleotides, respectively, while deletions are indicated by a triangle (▵). The lane numbers at the top of the autoradiogram correspond to the number of each ribozyme: 1, wild-type; 2, RzA78U,A79U; 3, Rz–U+79–80; 4, Rz–C19A,G81U,G80A; 5, Rz–U+76–77; 6, Rz–▵77; 7 is the original bimolecular construct (RzA–RzB); and, 8, the bimolecular construct including an abasic residue (Ab) in position 77 (RzB–Ab77). The positions of the substrate and product are indicated adjacent to the gel. The rate constants (kobs) for each ribozyme are indicated below the gel.