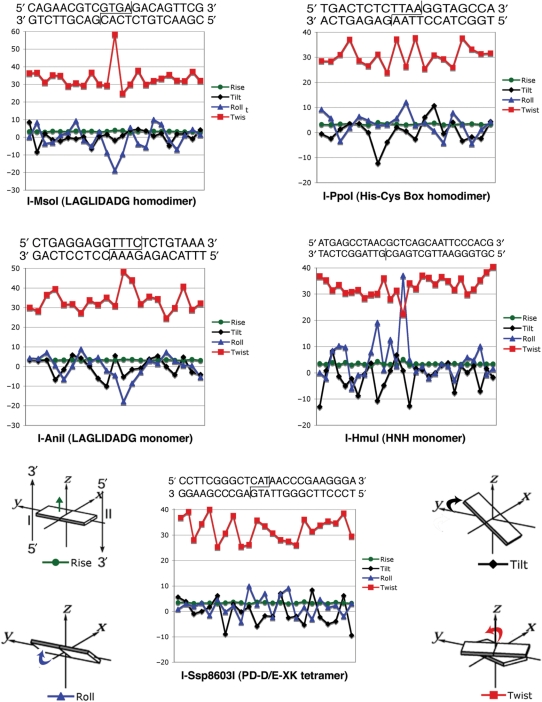
Figure 4.
DNA distortion induced by homing endonuclease binding. DNA bend parameters were quantitated using program ‘Readout’ (34), via a web-based server located at: http://gibk26.bse.kyutech.ac.jp/jouhou/readout/). The cognate target sequence, used in the individual crystal structures from which the bend parameters were calculated, are shown; cleavage sites are indicated. All endonucleases in this study except for I-HmuI cleave both strands to produce 3' overhangs; I-HmuI nicks the lower strand only (vertical line). The individual features of basepair steps distortion shown in the graphs (roll, tilt, twist and rise) are illustrated relative to a standard coordinate frame for double-stranded DNA.