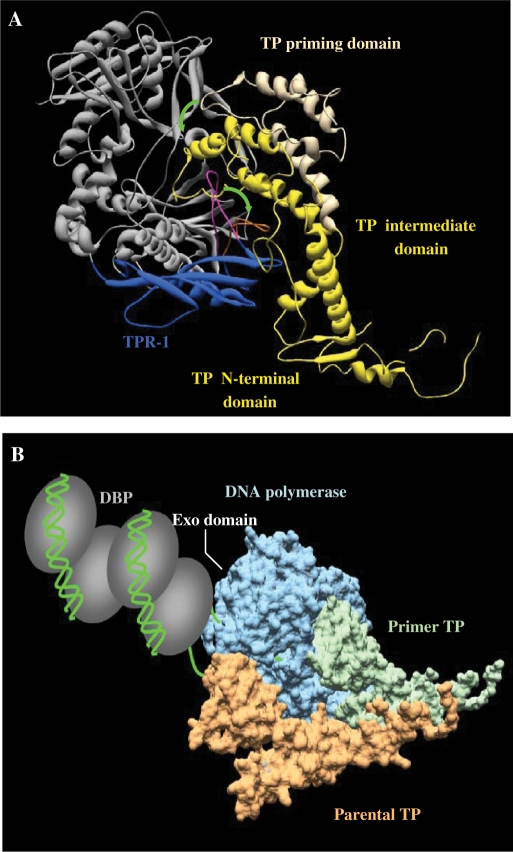
Figure 9.
Modelling of the temporal sequence of interactions that take place during initiation of TP-DNA replication. (A) Proposed conformational changes occurring during DNA polymerase/TP heterodimer formation. ϕ29 DNA polymerase is coloured in grey and the TPR-1 subdomain in dark blue. Flexible orientation of TPR-1 loop in the apoenzyme [PDB code 1XHX; (41)] and its stable and moved out structural conformation shown in the DNA polymerase/TP complex [PDB code 2EX3; (19)] is shown in magenta and orange, respectively. TP is coloured in yellow (TP priming domain in the proposed non productive orientation is indicated with light yellow). Green arrows indicate the suggested conformational changes of both, the DNA polymerase TPR-1 loop and the TP priming domain to allow the formation of a stable heterodimer. (B) Model of the DNA polymerase/TP recognition of replication origin. DNA polymerase molecular surface is coloured in blue, priming TP in green and parental TP in orange. The nucleoprotein complex at the replication origin, formed by DNA (depicted in green) and DBP (depicted as grey ovals) is mainly contacted by the DNA polymerase.