Fig. 1.
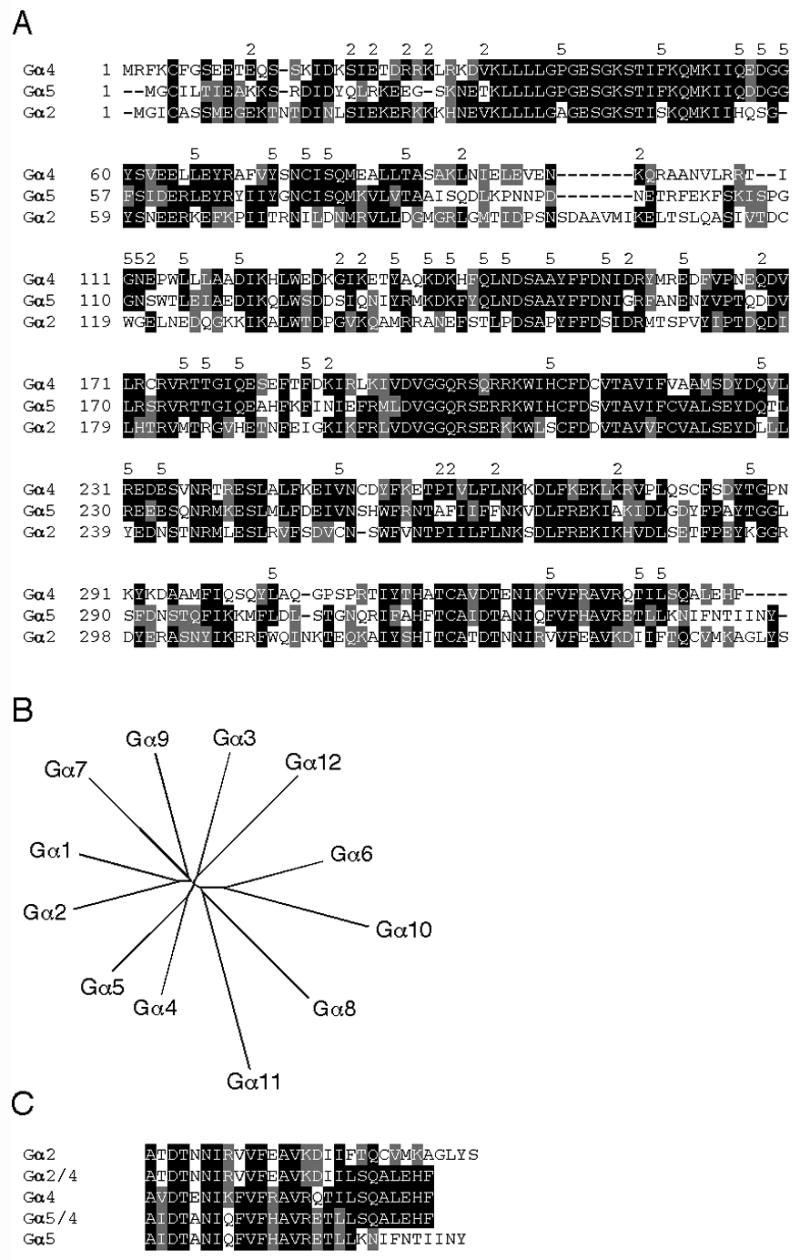
Comparison of Dictyostelium Gα subunits. (A) Alignment of Dictyostelium Gα4, Gα5 and Gα2 subunit amino acid sequences using ClustalW program and formatted using BOXSHADE program to highlight identical (black background) and similar residues (gray background). The number (2) above the aligned sequences indicates residues identical between the Gα4 and Gα2 but not Gα5 subunit. The number (5) above the aligned sequences indicates identity between the Gα4 and Gα5 but not Gα2 subunit. (B) Unrooted phylogenic tree of Dictyostelium Gα subunits generated at http://clustalw.genome.ad.jp/ by the neighbor-joining method from a ClustalW alignment. Sequence information was obtained using dictyBase at http://www.dictybase.org/. (C) Alignment of wild-type and chimeric Gα subunit carboxyl terminal amino acid sequences formatted using the BOXSHADE program as described above.
