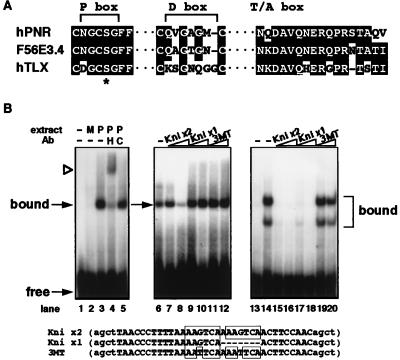
Figure 2.
(A) Alignment of the amino acid residues in the P box, D box and T/A box regions of the DBDs between human PNR, C. elegans F56E3.4 and human TLX. Conserved residues are shaded. Gaps are indicated by a dashed line. ∗ indicates the position of the serine residue in the P boxes of these 3 receptors that replaces an otherwise strictly conserved lysine. (B) In vitro DNA binding specificity by PNR protein. Lanes 1–5 show DNA-binding patterns of COS cell-expressed HA-tagged PNR protein to Kni x2 probe. Lane 1 is probe only, and mock cell extract was added in lane 2. Lane 3 shows a specific protein–DNA complex formed when cell extract containing HA-hPNR is added. This band is supershifted by addition of antibody against the HA tag (lane 4) but is not affected by nonspecific antibody (lane 5). Lanes 6–12 show that PNR bound to labeled Kni x2 probe can be blocked by competition by addition of excess Kni x2 cold probes (lane 8), but not by Kni x1, which is based on the TLL binding site identified in the D. melanogaster knirps gene (17), or 3MT probes (lanes 9–12). Competitor probes are indicated above, with 10-fold excess added in lanes 7, 9, and 11 and 50-fold excess in lanes 8, 10, and 12. Lanes 13–20 show that COS cell-expressed chicken TLX protein forms two distinct complexes on the Kni x2 probe (lane 14), both of which can be efficiently blocked by competition by either Kni x2 or Kni x1, but not the mutated 3MT probe.