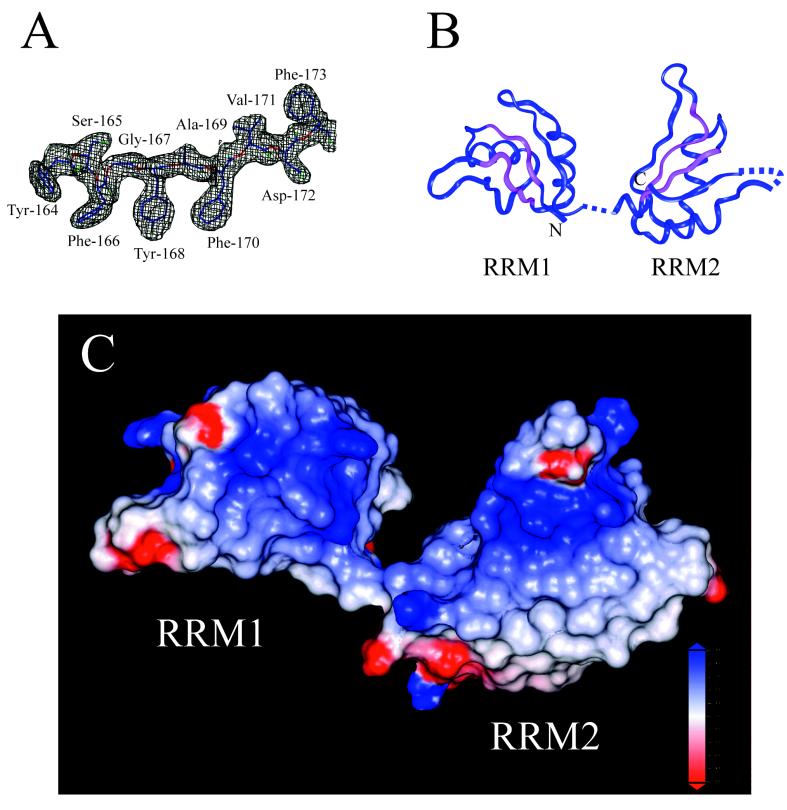
Figure 1.
Structure of the SXL RRM domains. (A) Sigma-A-weighted, 2Fo − Fc electron density map contoured at 1σ illustrates the quality of the map. Residues shown are Tyr-164–Phe-173. (B) Ribbon diagram of SXL RRM1+2. Residues left out of the model for lack of interpretable electron density include 111–123, 204–205 (interdomain linker), 276–282 (α2-β4 loop of RRM2), and 290–294 (C terminus). RNP-1 and RNP-2 sequences are colored in magenta. (C) Electrostatic surface potential of SXLl+2 calculated with grasp (blue = +5 kT > white = 0 kT > red = −5 kT). The absence of interdomain contacts in the SXL RRMl+2 crystal structure is evident. Based on the structures of the RNA complexes of UlA and U2B′′/U2A′, the most positive regions are likely RNA binding surfaces.