Summary
Tumor necrosis factor alpha (TNF-α), a pro-inflammatory cytokine, plays a key role in the pathogenesis of many inflammatory diseases, including alcoholic liver disease. In the liver, Kupffer cells are the primary source of the cytokine. Obliteration of Kupffer cells or neutralization of TNF-α by anti-TNF-α antibody or by an antisense oligonucleotide prevents ethanol-mediated liver damage. In this study, we report the identification of yet another highly efficacious gene-silencing molecule, the short interfering RNA (siRNA), SSL3, against TNF-α. The efficacies of various siRNA duplexes were tested against TNF-α mRNA in primary cultures of rat Kupffer cells. SSL3 (25 nM) inhibited lipopolysaccharide (LPS)-induced secretion of TNF-α by 55% (p<0.005) with a proportionate reduction in TNF-α mRNA; the inhibitory effect lasted for at least 96 h. Four nucleotide mismatches to SSL3 completely abolished the inhibitory effects of SSL3, suggesting the sequence specificity of the siRNA. Further, the in vivo efficacy of SSL3 was assessed following the i.v. administration of two doses (140 μg/kg body weight/day for two days) of liposome-encapsulated SSL3. The LPS-induced TNF-α secretion was inhibited by > 60% (p < 0.05) by SSL3 pre-treatment. These data demonstrate the identification of an siRNA against TNF-α, which, as a liposomal formulation, has therapeutic potential in the treatment of inflammatory diseases mediated by TNF-α.
Keywords: TNF alpha, siRNA, Kupffer cells, lipopolysaccharide, liposomes
Introduction
In response to physiologically challenging conditions, such as endotoxemia or immune reactions, macrophages release excessive amounts of cytokines, interleukins and prostanoids, often resulting in organ damage. In the liver, Kupffer cells, the resident macrophages, play an important role in the pathogenesis of liver injury. It has been demonstrated that obliteration of Kupffer cells prior to their activation by hepatotoxins prevents liver damage [1-3]. Tumor necrosis factor alpha (TNF-α), a cytokine produced by many cell types, including macrophages, monocytes, lymphocytes and fibroblasts, is a growth promoter under normal physiological conditions. It exhibits pleiotropic effects on various cell types [4]. However, excessive amounts of the cytokine are released into circulation in response to infection, inflammation and other environmental insults, often resulting in tissue injury [5]. In the liver, Kupffer cells are the major producers of TNF-α following exposure to lipopolysaccharide (LPS), the bacterial endotoxin [6]. An overproduction of TNF-α has been associated with the development of alcoholic liver injury [7-9], rheumatoid arthritis [10], inflammatory bowel disease [11] and septic shock [12]. Antibodies against TNF-α, neutralize the effects of TNF-α in conditions such as ischemia reperfusion [13] and experimental liver damage induced by chronic alcohol consumption [14]. From the above, it is concluded that any approach to mitigate the excessive production of pro-inflammatory cytokines, such as TNF-α, would be of therapeutic value. Further, because they are the major source of cytokines in the liver, targeting Kupffer cells would have to be the primary objective in any intervention.
Recent studies from our laboratory have shown that antisense oligodeoxynucleotides (ODNs) directed against TNF-α mRNA can effectively prevent ethanol-induced liver damage [15]. Although theoretically, antisense ODNs can be used to target any of the disease-causing genes, in practice, there are limitations to its application due to off-target effects [15,16]. Against that background, the discovery of RNA interference (RNAi) is of significance. The discovery that the administration of short interfering RNA (siRNA) can activate an endogenous cellular process for gene-specific silencing has led to an explosion of interest in this field [17]. Under appropriate conditions, short (21-23 nucleotides) double-stranded RNA (dsRNA) molecules are generated from a double-stranded parent molecule by an endonuclease called the ‘Dicer’, an RNAse III enzyme [18]. In turn, the short-dsRNAs, the siRNAs, are incorporated into an enzyme complex, the RNA-induced silencing complex (RISC), which unwinds the double strand and is guided by the antisense strand to the homologous mRNA for degradation [18-20]. Earlier studies by Brummelkamp et al.,[21] and Bertrand et al.,[22] suggest that siRNAs are highly sequence specific and are much more sensitive compared to the antisense oligonucleotides. The extraordinary success in gene-silencing realized by the RNAi system has also led to several in vivo applications. A comprehensive review on the in vivo application of siRNAs has recently appeared [23].
The commonly used in vivo delivery systems include, adenoviral vectors coding for hairpin RNA molecules (shRNA) that generate siRNA in the cell, cationic lipids, hydrodynamic injection, electroporation and also the ‘naked’ form of administration [23]. While the choice of the delivery system depends upon the target, it is generally observed that delivery of oligonucleotides in the ‘naked’ form is the least efficient one. In the studies described here, we report the identification of an siRNA duplex that was effective against rat TNF-α mRNA, and its successful delivery in vivo using an anionic liposomal delivery system.
Materials and Methods
Animals
All experiments were carried out in barrier maintained male Sprague Dawley rats purchased from Harlan (Indianapolis, IN). Animals were acclimatized for at least 1 week following their arrival in our facility. The body weights of the animals used ranged from 275-325 g. All animals were maintained on lab-chow until used. The experimental protocol was approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University, Philadelphia, which is an AAALAC-accredited facility.
Chemicals
Cholesterol and cholesteryl hemisuccinate (CHEMS) were purchased from Sigma-Aldrich (St. Louis, MO). Phosphatidyl ethanolamine (PE) (transphosphatidylated from egg lecithin) was purchased from Avanti Polar Lipids Inc. (Alabaster, AL). siRNA duplexes targeted to rat TNF-α mRNA were synthesized (Dharmacon Inc., Lafayette, CO) either as individual siRNA duplexes or as a mixture of four siRNA duplexes, referred to as the ‘smart pool’siRNA (SM-siRNA). The sequences of the individual siRNAs are follows: SSL1, Sense - 5’- G-C-A-G-A-U-G-G-G-C-U-G-U-A-C-C-U-U-A-U-U-3’ Antisense - 5’-P-U-A-A-G-G-U-A-C-A-G-C-C-C-A-U-C-U-g-C-U-U-3’ ; SSL2, Sense 5’-C-A-A-C-C-U-G-C-C-C-A-A-G-U-A-C-U-U-A-U-U-3’, Antisense 5’-P-U-A-A-G-U-A-C-U-U-G-G-G-C-A-G-G-U-U-G-U-U-3’; SSL3, Sense 5’-U-C-U-C-A-A-A-A-C-U-C-G-A-G-U-G-A-C-A-U-U-3’, Antisense 5’-P- U-G-U-C-A-C-U-C-G-A-G-U-U-U-U-G-A-G-A-U-U-3’; SSL4, Sense 5’-C-C-A-U-U-U-C-A-U-A-C-C-A-G-G-A-G-A-A-U-U-3’, Antisense 5’-P U-U-C-U-C-C-U-G-G-U-A-U-G-A-A-A-U-G-G-U-U-3’. The sequence of the non-silencing negative control (N-Control) siRNA is as follows: N-Control, Sense, 5’-U-AG-C-G-A-C-U-A-A-A-C-A-C-A-U-C-A-A-U-U-3’, Antisense 5’-P- U-U-g-A-U-G-U-G-U-U-U-A-G-U-C-G-C-U-A-U-U-3’. Transfection reagent, RNAiFect™ (a cationic liposome delivery reagent), RNA extraction kit, RNeasy® Mini Kit and the reverse transcription kit, Qunatifect® were all purchased from Qiagen Inc. (Valencia, CA). Products for the Real Time PCR were all purchased from Applied Biosystems (Foster City, CA); Taqman Universal PCR master mix (Cat # 4304437), forward and reverse primers for rat TNF-α (Cat # Rn00562055_m1) and the forward and reverse primers for rat β-actin (Cat # Rn 00667869_m1). All other chemicals used were of reagent grade, purchased either from Sigma-Aldrich (St. Louis, MO) or from Fisher Scientific (Pittsburgh, PA).
Preparation of Kupffer cells
Kupffer cells were prepared as previously described by Ponnappa et al [24], which is a modification of the procedure originally described by Bautista et al., [25]. Briefly, following anesthesia (Ketamine plus Xylazine), the liver was pre-perfused in situ with Ca2+ - and Mg2+ -free Hank’s Balanced Salt Solution (HBSS, GIBCO-BRL, Gaithesberg, MD) for 20 min followed by a sequential perfusion with pronase (Sigma Cat # P-5147) and collagenase (Sigma Cat # C-5138) in HBSS (Ca2+ - and Mg2+ -reconstituted). All the solutions used in the perfusion were maintained at 37°C. After the perfusion, the liver was carefully cut into small pieces and placed in 90 ml of HBSS containing a mixture of collagenase and DNase (Sigma Cat # D-4527). The contents were shaken in an Orbital Shaker (Forma Scientific) at 250 rpm and 37°C for 15 min. The digested liver suspension was passed through a mesh to separate the dissociated cells from the undigested tissue. The heavier hepatocytes were separated from the non-parenchymal cells by centrifugation at 50 g for 2 min. The pellet was washed three times with HBSS for maximum recovery of the non-parenchymal cells into the supernatant. The pooled supernatant was further centrifuged at 430 g for 6 min to pellet the non-parenchymal cells, washed twice and finally resuspended in 10 ml of in Gey’s Balanced Salt Solution (GBSS) for centrifugal elutriation. Kupffer cells were separated from other cell-types by centrifugal elutriation (J2-MC centrifuge, JE-6B elutriator rotor, Beckman Coulter, Inc., Fullerton, CA). Purity of the ‘Kupffer cell’ fraction (>85% pure) was routinely monitored by peroxidase staining. Kupffer cells stain brown and endothelial cells stain blue. The Kupffer cell fraction was further washed and resuspended in PBS. Viability, as determined by trypan blue exclusion, was generally greater than 95%.
Tissue culture
Freshly isolated Kupffer cells were routinely plated in 12-well plates at a density of 1-1.2 × 106 cells/well in a culture medium consisting of 1640 RPMI supplemented with 10% fetal bovine serum (FBS), 30 mM Hepes buffer, 1 mM pyruvate and antibiotics. Cells were cultured at 37°C in 5% CO2 atmosphere and allowed to attach for 1 h, after which, the medium was changed. Routinely, Kupffer cells were cultured for 4-6 days before transfection with siRNAs.
Real-time RT-PCR
Total RNA was isolated from Kupffer cells using RNeasy® Mini Kit. After genomic DNA elimination, RNA was reverse-transcribed using QuantiTect® Reverse Transcription Kit according to manufacture’s instructions. For real-time PCR, 10 ng of the reverse transcriptase product was used for amplification of TNF-α or β-actin genes using respective primers and TaqMan MGB probe (6-FAM dye-labeled) together with TaqMan Universal PCR Mix. The plate was run on the ABI 7000 Sequence Detection System. Standard curves were generated and the relative quantity of target gene mRNA in each sample was normalized to β-actin mRNA. All reactions were done in triplicate.
Preparation of siRNA-encapsulated liposomes
Liposome-encapsulated siRNA was prepared by the reverse phase method originally described by Szoka and Papahadjopolous [26] with modifications as reported by Ponnappa et al., for the preparation of antisense oligonucleotides [27]. Briefly, a mixture (25 mg) of lipids (PE, CHEMS and cholesterol in a molar ratio of 7:4:2,) was dissolved in 4.5 ml of chloroform. Two mg of SSL3 was dissolved in 0.4 ml of Tris-EDTA (TE) buffer (10 mM Tris, 1 mM EDTA, pH 8) and diluted to 1.5 ml with a hypotonic buffer (made up of 1:9 diluted phosphate buffered saline (PBS) supplemented with 25 mM sodium phosphate, pH 7.4). The aqueous siRNA solution was added to the lipid mixture in chloroform and sonicated in a bath-type sonicator for 5 min. The organic solvent was evaporated at room temperature using a rotary-type evaporator (BUCHI Rotovapor R-134, Flawil, Switzerland). The resultant liposomal suspension was diluted with the hypotonic buffer and centrifuged at 100,000 × g for 45 min to separate the liposomes from the medium. The liposomal pellet was washed twice with modified PBS (PBS supplemented with 25 mM phosphate buffer, pH 7.4) and resuspended in a volume not exceeding 1 ml of modified PBS. Empty liposomes, used as the placebo in the in vivo studies, were prepared using the same proportion of buffers and lipid mixture but without the siRNA. The concentration of encapsulated siRNA was determined spectrophotometrically at 260 nm, after extracting siRNA from an aliquot of the liposomal preparation. Routinely, the amount of siRNA encapsulated by the liposomes ranged from 10-12% of the total siRNA taken for encapsulation.
In vivo studies
The in vivo efficacy of the siRNA, SSL3, was determined using the liposomal delivery method that was successfully used in our laboratory for antisense oligonucleotides [15, 27]. Briefly, preparations of liposome-encapsulated SSL3 were injected intravenously (in a volume of PBS not exceeding 1 ml) into the animal by the tail vein route. Rats were injected with 0.14 mg/kg body weight of SSL3 each day for two successive days. On the fourth day, the animals were injected with LPS (50 μg/Kg body weight, i.v.) to induce TNF-α secretion and sacrificed 90 min later. Venous blood was collected and allowed to coagulate at room temperature. Following centrifugation, serum was collected and stored at -80°C until TNF-α levels were determined by ELISA. Previous studies have shown that it required 48 h post intravenous injection of the liposomes for maximal efficacy, most likely due to the slow release of the liposomal contents into the cytosol [27] Animals injected with empty liposomes were treated as ‘control’. In all of the experiments, the concentration of the liposomal lipid injected was maintained below 16 mg/kg body weight to avoid non-specific effects on LPS-induced TNF-α secretion [27].
TNF-α assay
TNF-α was assayed by ELISA using a kit (Cat # 88-7340-88) from eBioscience (San Diego, CA) as per manufacturer’s specifications. Serum or cell culture media, treated with LPS, were diluted appropriately to make sure that TNF-α values were within the range of the standard curve.
Protein assay
Cells were solubilized in 0.5 N NaOH and assayed after dilution for total protein content using Micro BCA™ protein assay kit (Kit # 23235, Pierce, Rockford, IL) using bovine serum albumin as the standard.
Statistical analysis
Where indicated, values are expressed as the means ± S.E. of (n) determinations. Statistical differences were evaluated between two samples by Student’s t-Test using the Microsoft Excel program. p values of < 0.05 were considered statistically significant.
Results
In vitro studies
Efficacy of anti-TNF-α siRNA
Although algorithms currently available for designing siRNA are evolving to be highly efficient, it is necessary to test the efficacy of more than just one siRNA construct, since it is unlikely that all targets would be equally susceptible for RISC-mediated hydrolysis. Therefore, the strategy for identifying an efficacious siRNA against TNF-α mRNA started with testing the efficacy of a mixture of four siRNA constructs, referred to as ‘smart pool-siRNA (SM-siRNA, Dharmacon Inc.,). Following transfection, the efficacy of SM-siRNA was determined by monitoring the LPS-induced secretion of TNF-α, in vitro, in primary cultures of rat Kupffer cells. Cells treated with a negative control siRNA (NC-siRNA) served as one of the controls. Additional controls included those that were exposed to PBS and the transfection reagent, RNAifect™. As shown in Fig. 1, at 100 nM, SM-siRNA inhibited LPS-induced TNF-α production in Kupffer cells by greater than 70% (p < 0.005) compared to cells treated with transfection reagent alone. There was no significant difference in the TNF-α secretion between the control groups.
Figure 1. Effect of SM-siRNA on TNF-α secretion in Kupffer cells.
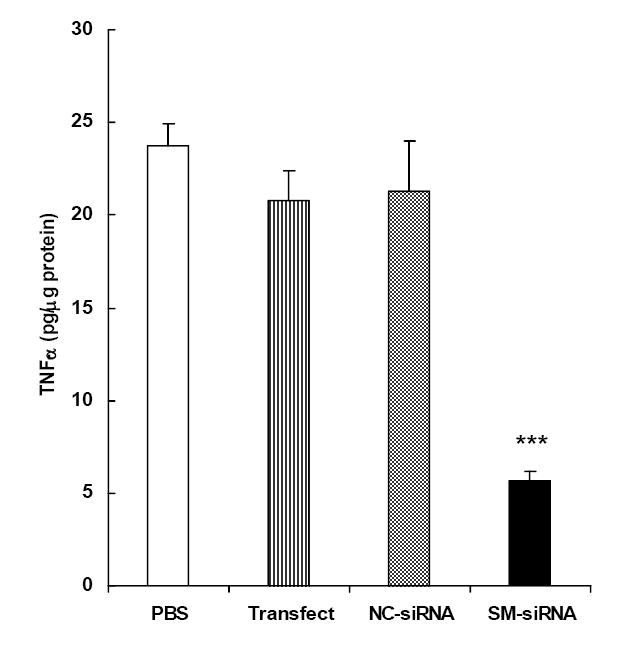
Freshly isolated rat Kupffer cells from male Sprague-Dawley rats were cultured in 12 well-plates for 4-6 days as described in Methods. Cells were transfected (RNAiFect™) with a mixture of four siRNAs (SM-siRNA,) targeted to rat TNF-α mRNA. Cells exposed to negative control siRNA (NC-siRNA), transfection reagent (Transfect) or PBS, were included as controls. siRNAs were used at a concentration of 100 nM. Twenty four hours after the transfection, cells were exposed to LPS (0.1 μg/ml) for 2 h to induce TNF-α secretion. TNF-α secreted into the medium was assayed by ELISA. Data points are the means ± S.E. (n=4). *** p < 0.005 compared to the control group treated with transfection reagent alone.
Since SM-siRNA was highly effective in inhibiting TNF-α secretion, it was logical to identify the individual siRNA component(s) responsible for the inhibitory effect in SM-siRNA. The efficacies of the individual components (SSL1, SSL2, SSL3 and SSL4) of the siRNA pool were tested in primary cultures of rat Kupffer cells as described above. As shown in figure 2, among the four siRNAs, only SSL1 and SSL3 showed inhibitions of 30% (p < 0.05) and 70% (p < 0.005) respectively. Thus, SSL3 emerged as the most efficacious siRNA against LPS-induced TNF-α secretion in Kupffer cells.
Figure 2. Effect of individual siRNAs on TNF-α secretion in Kupffer cells.
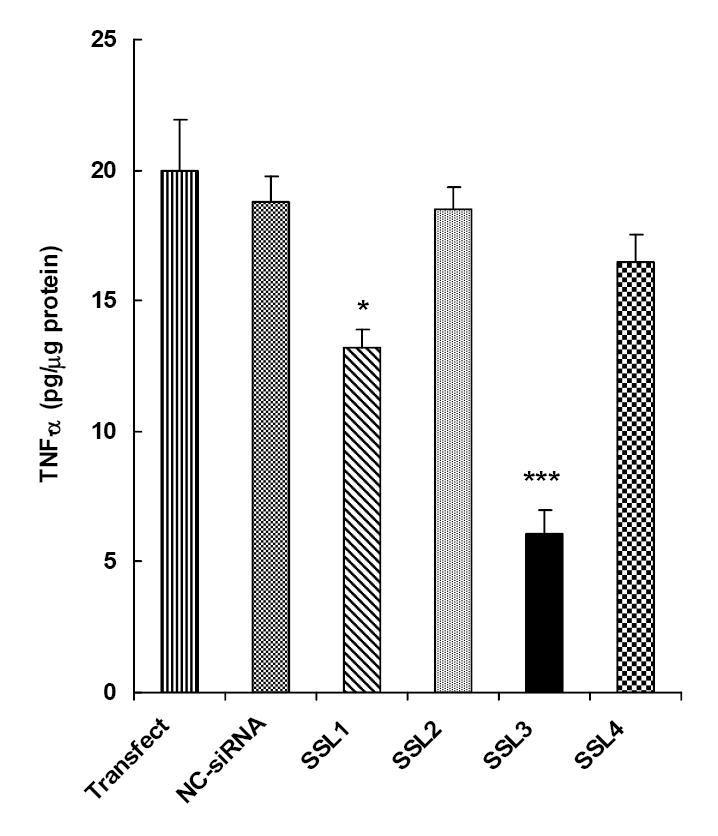
Primary cultures of rat Kupffer cells were transfected with each of the four (SSL1, SSL2, SSL3 and SSL4) components of the SM-siRNA pool described in Fig. 1 siRNAs were used at a concentration of 100 nM. Other conditions were similar to that described in legend to Fig. 1. The data points are the means ± S.E. (n=4). *p < 0.05, *** p < 0.005 compared to the control group treated with transfection reagent alone.
Dose-response studies
In the studies described above (Figs 1 and 2), the efficacies of SM-siRNA and the individual siRNAs were tested at 100 nM, a recommended starting concentration for newly synthesized siRNAs. However, many siRNA constructs have been reported to demonstrate high degree of sensitivity at lower concentrations. Therefore, we tested the dose-dependent effects of SSL3 in the range of 10-100 nM. As shown in figure 3, SSL3 was highly effective (p <0.005) at 25 nM, inhibiting TNF-α secretion by 55% compared to those cells treated with NC-siRNA. Under identical conditions, at 50 and 100 nM, inhibitions were marginally higher at 60% and 65% respectively. Therefore, in the subsequent studies, we used SSL3 at the lower efficacious concentration of 25 nM. At 10 nM, a lower but significant inhibition (30%, p < 0.05) of TNF-α secretion by SSL3 was also observed.
Figure 3. Effect of siRNA SSL3, on TNF-α secretion in Kupffer cells: dose-response studies.
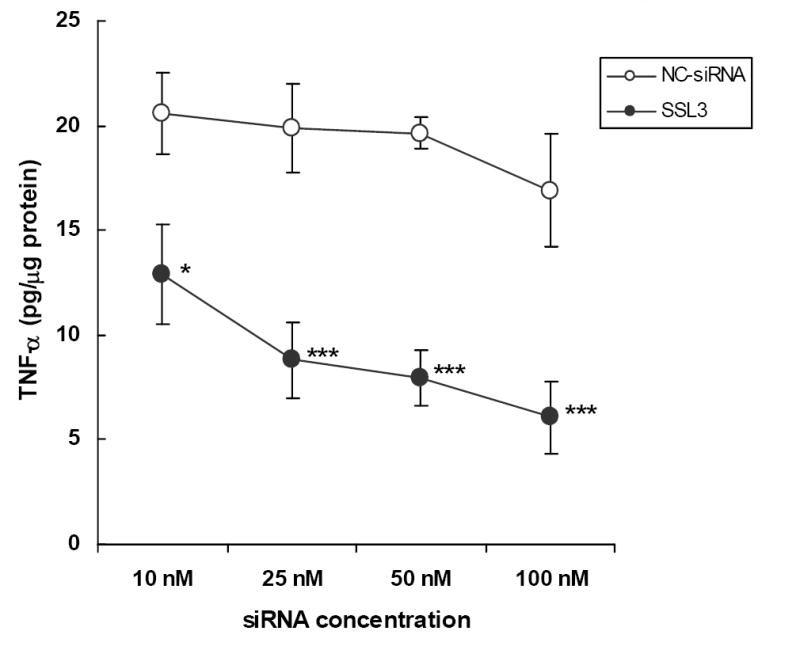
Primary cultures of rat Kupffer cells were transfected with various concentrations (10-100 nM) of SSL3. The efficacy of the siRNA was determined following LPS stimulation as described in legend to Fig. 1. Data points indicate TNF-α values which are the means ± S.E. (n=4). * p < 0.05, *** p < 0.005 compared to controls treated with NC-siRNA.
Sequence specificity
Studies on siRNAs are generally carried out on the assumption that the short, double-stranded RNAs induce inhibitory effects by targeting specific nucleotide sequences on mRNAs and degrade the same by RISC-mediated mechanisms [18-20]. To confirm the same, we introduced four mismatches (SSLM) to SSL3, flanking the mid region of the siRNA (SSLM, sense, 5’- U-C-U-C-A-A-A-G-U-A-U-G-A-G-U-G-A-C-A-U-U-3’ and antisense, 5’- P-U-G-U-C-A-C-U-C-A-U-A-C-U-U-U-G-A-G-A-U-U-3). Comparison of efficacy data between NC-siRNA, SSL3 and SSLM suggest that alteration of the nucleotide sequence completely abolishes the inhibitory effects of SSL3 on LPS-induced TNF-α secretion, confirming the sequence-specificity of SSL3 (Fig. 4).
Figure 4. Sequence specificity of SSL3.
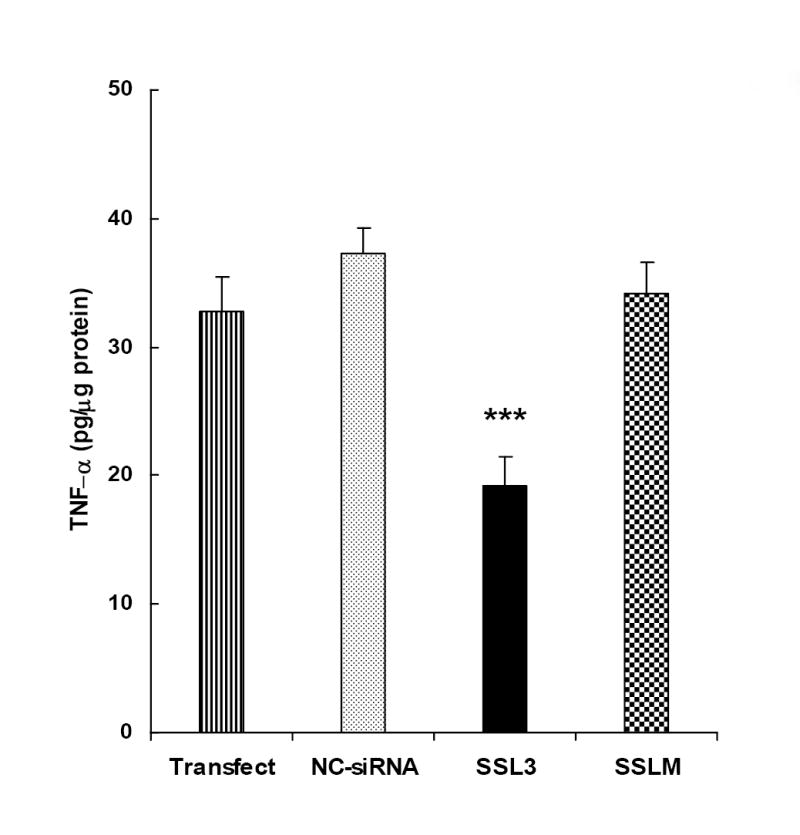
The sequence specificity of the siRNA, SSL3 was assessed by comparing its efficacies of SSL3 with that of SSLM, an siRNA, in which, four bases in the mid region of SSL3 were replaced. The sense sequences of SSL3 and SSLM are as follows: SSL3, 5’- U-C-U-C-A-A-A-A-C-U-C-G-A-G-U-G-A-C-A-U-U-3’; SSLM, 5’- U-C-U-C-A-A-A-G-U-A-U-G-A-G-U-G-A-C-A-U-U-3’. Cultured rat Kupffer cells were transfected with 25 nM of each of the siRNAs including NC-siRNA, and their efficacies were determined 24 h post transfection as described in legend to Fig. 1. Bars indicate values of TNF-α secretions which are the means ± S.E. of 4-6 determinations. ***p < 0.005 between SSL3 and NC-siRNA-treated cells.
TNF-α mRNA levels
The gene-silencing effect of SSL3 on TNF-α mRNA was further confirmed by real-time-PCR. Similar to the inhibitory effects on TNF-α secretion (Fig. 3), transfection of Kupffer cells with 25 nM SSL3 reduced TNF-α mRNA levels by greater than 50% compared to NC-siRNA or SSLM (Fig. 5). The data suggest that SSL3-mediated inhibition of TNF-α secretion in Kupffer cells is brought about by the breakdown of TNF-α mRNA, confirming the mechanism of RNAi.
Figure 5. Determination of TNF-α mRNA levels by Real-time RT-PCR.
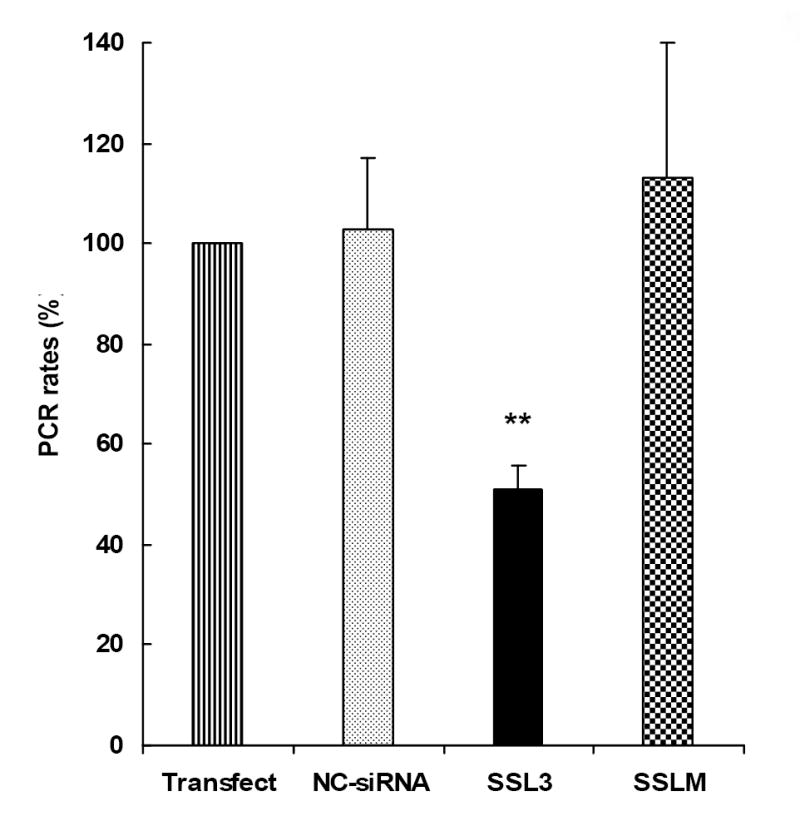
Freshly isolated Kupffer cells were cultured for 6 days followed by transfection with 25 nM siRNAs, NC-siRNA and SSL3. Cells treated with transfection reagent and NC-siRNA were included as controls. Twenty four hours after the transfection, cells were exposed to LPS for 1 h and the total RNA was extracted and reverstranscribed to cDNAs. 10 ng (cDNA) aliquots of each sample was taken for the determination of quantitative real-time PCR as detailed in Methods. The PCR rates of TNF-α mRNA synthesis in various treatment groups were normalized to the rate of synthesis of β-actin, included as the endogenous control. Data were plotted as percent PCR value of 100 to samples treated with the transfection reagent alone. Bars indicate PCR values which are the means + S.E. of n=3 determinations. **p < 0.01 between SSL3 and Transfect control.
Time-course studies
The temporal effects of SSL3 against TNF-α was determined by exposing Kupffer cells to the siRNA (25 μM) for up to 96 h, after which, they were stimulated with LPS to induce TNF-α secretion. Data in figure 6 show that the inhibitory (50-70%, p < 0.01-< 0.001) effects of SSL3 on TNF-α production persisted for at least 4 days, the maximal duration studied.
Figure 6. Effect of siRNA, SSL3 on TNF-α secretion: Time-course studies.
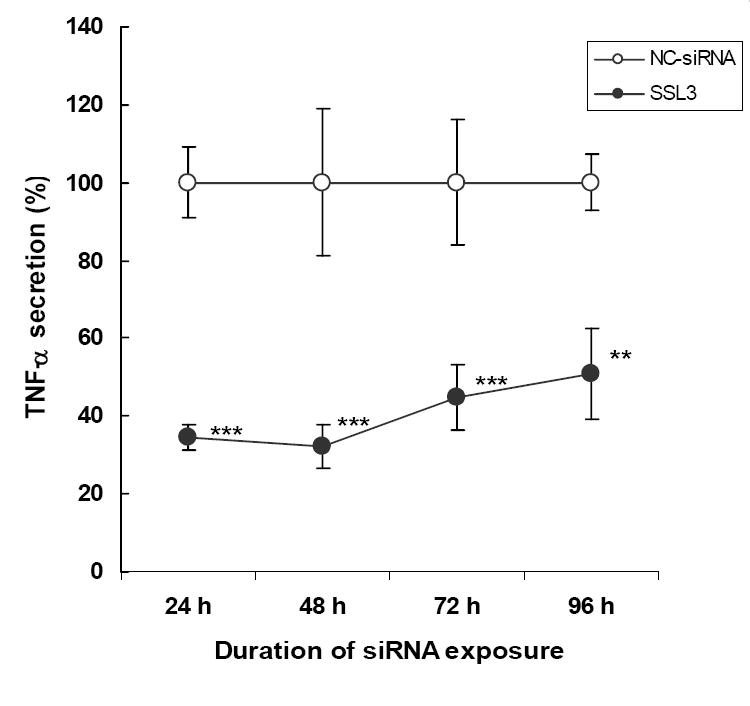
Freshly isolated Kupffer cells were cultured for 6 days followed by transfection with 25 nM of the siRNAs. At 24, 48, 72 and 96 h post transfection, TNF-α secretion was induced by LPS for 2 h. Data points indicate TNF-α values which are the means ± S.E. (n=7-8). TNF-α secretion in cells treated with SSL3 was expressed as percent of secretions by those samples treated with NC-siRNA, which had no inhibitory effect on LPS-induced TNF-α secretion (Figs 1-5). **p < 0.01, ***p < 0.005.
In vivo studies
In vivo efficacy of SSL3
The in vivo efficacy of SSL3 was determined using the liposomal delivery system already established in our laboratory for targeting ODNs to Kupffer cells in vivo [15,27]. Rats were intravenously injected either with liposome-encapsulated siRNA, SSL3 or with empty liposomes, used as the control. Conforming to the established protocol from our previous studies (with S-ODNs) [27], rats were injected with liposome-encapsulated SSL3 (0.14 mg/kg body wt/day) for 2 days and challenged with LPS on the fourth day. Data presented in figure 7 show that TNF-α secretion was inhibited by greater than 60% (p <0.05) by SSL3-liposomes compared to the empty liposome-injected control group. We have already reported that empty liposomes per se, when used at lipid concentrations (15-17 mg/kg body wt.) do not affect LPS-induced TNF-α secretion [27].
Figure 7. In vivo efficacy of SSL3.
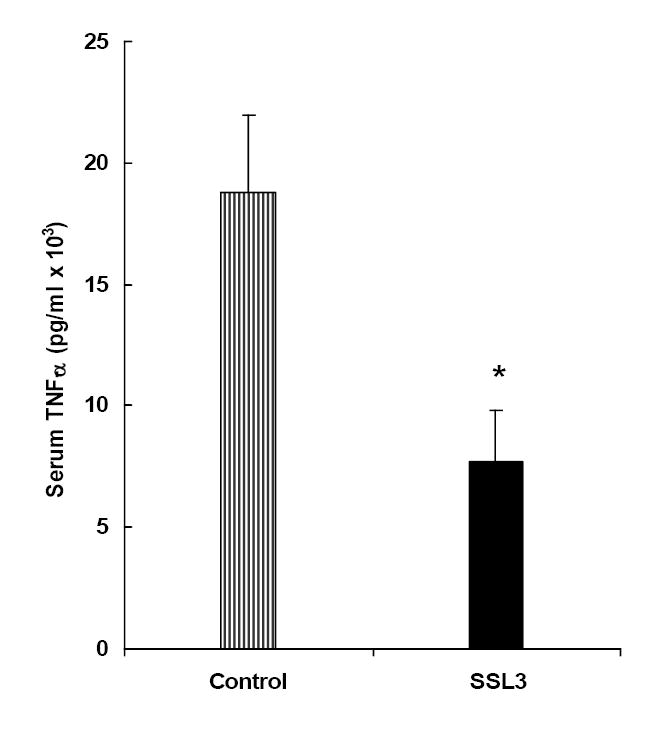
The in vivo efficacy of SSL3, the 21-mer anti-TNF-α siRNA, was determined following its encapsulation in pH-sensitive liposomes. Male S.D rats were injected with liposome-encapsulated SSL3 (0.14 mg/kg body wt) by tail vein, daily for two days. On the fourth day, TNF-α secretion was induced by LPS (50 μg/kg body wt) and blood was collected at 90 min post LPS for the determination of serum levels of TNF-α. Rats injected with an equal dose of “empty liposomes” were treated as the control group. Bars indicate serum TNF-α values which are the means ± S.E. from n=4 rats per group. *p < 0.05.
Discussion
One of the challenges in drug development is to achieve high specificity and least toxicity. Antisense technology was developed with the idea that short nucleotides of certain sequence are found only once in the entire genome and therefore, gene silencing with specific ODNs could be highly specific. However, issues such as antisense instability in plasma and off-target effects due to back-bone modifications etc. have hampered the realization of its full potential. Nevertheless, under a defined set of circumstances, it has been possible to harness the potential of ODNs. For example, macrophages are the primary source of TNF-α [6]. Earlier, we demonstrated that liposomes, in the size range above 200 nm in diameter, primarily target Kupffer cells and splenic macrophages [24] and therefore, it is possible to selectively target a population of cells, such as the macrophages, which constitute a small percentage (< 0.2%) of the body weight. In such a case, off-target effects, if any, are mostly confined to a very small population of cells. Using an anti-TNF-α S-ODN and the liposomal delivery system, we have demonstrated that it was possible to inhibit > 70% of LPS-induced TNF-α secretion in vivo at relatively low concentration (1-2 mg/kg body weight) of the antisense; for a similar effect, it required a 5-10-fold higher concentration of the S-ODN if injected in the free form [15]. Thus, our earlier studies demonstrated that liposomal delivery system can indeed be a viable option for the in vivo targeting of oligonucleotides to Kupffer cells and other macrophages.
The discovery of RNA interference, with the associated high sensitivities of siRNAs as gene-silencing agents, has provided newer opportunities in the treatment of many diseases [23]. As indicated earlier, TNF-α plays a pivotal role in the pathogenesis of many inflammatory diseases [7-12]. Having successfully used the liposomal delivery system to down-regulate TNF-α using antisense oligonucleotides, it was our objective to identify highly effective siRNA duplexes against TNF-α so as to be able to achieve in vivo efficacy at as low a concentration of the nucleotide as possible. Studies were initiated by testing the efficacy of a pool of four siRNA constructs against rat TNF-α mRNA. As expected, not all of the constructs were equally effective, suggesting that not all the domains on the mRNA transcript are viable targets for siRNA binding. One of the constructs, SSL3 was the most potent one, which exhibited 65-70% inhibition of LPS-induced TNF-α secretion in primary cultures of rat Kupffer cells. The absence of significant inhibition with NC-siRNA as well as with the mismatched SSL3 (SSLM), suggested that SSL3-mediated effects on TNF-α was sequence specific. Reductions in efficacy was also observed when two or three mismatches were incorporated into SSL3 (data not shown). Further confirmation of RNAi-mediated mechanism of action of SSL3 came from real-time -PCR studies, in which, the reduction in mRNA levels paralleled the extent of inhibition of TNF-α secretion in Kupffer cells. Time-course studies in primary cultures of rat Kupffer cells showed that the inhibitory effect of SSL3 remained relatively constant for at least 4 days, the longest duration studied. This observation is in line with those of others [22,28], who have reported longer half-lives (than S-ODNs) for siRNAs in non-dividing cells. Taken together, our data confirm the identification of an anti-TNF-α siRNA construct, which can be potentially used in the treatment of inflammatory diseases.
Although it is often possible to identify highly efficacious gene-silencing agents in vitro using cell culture systems, their therapeutic values lie in our ability to demonstrate their in vivo efficacy. Similar to other drugs, the major challenge facing siRNA therapeutics is in their in vivo efficacy, which requires an efficient delivery system. In most cases, siRNAs are delivered in the ‘naked’ form, which has the inherent problem of being subject to degradation in plasma, similar to antisense ODNs. To overcome such a problem, several chemical modifications to siRNAs have been attempted but off-target effects are a major concern [23]. Against that background, an ideal delivery system was considered to be the one, in which, an siRNA could be delivered to the target cells without interference from plasma components. Since we have already demonstrated that the liposomal formulation used in the current study protects oligonucleotides from degradation in plasma [27], we considered the pH-sensitive anionic liposomes as an ideal delivery vehicle for targeting siRNA to Kupffer cells and other macrophages in vivo. The in vivo efficacy was assessed by measuring LPS-induced serum levels of TNF-α following delivery of liposome-encapsulated SSL3. We observed that SSL3 inhibited LPS-induced TNF-α secretion by 60% (Fig. 7), which is similar to the effect exhibited by the anti-TNF-α S-ODN used in an earlier study but at ten times lower concentration [15]. These data clearly suggest that siRNAs can be successfully used in vivo with a high degree of efficacy. It is also to be pointed out that in other models using the ‘naked’ form, it was necessary to administer multiple doses of a much higher siRNA concentration for in vivo efficacy [23].
Although the primary focus our studies has been to target Kupffer cells, it is to be pointed out that spleen, by way of splenic macrophages, also has a high capacity to produce TNF-α, in response to LPS stimulation [15]. We have observed that intravenous administration of an anti-TNF-α antisense oligonucleotide inhibits LPS-induced TNF-α secretion by similar levels in plasma, liver and spleen [15]. More interestingly, splenectomy completely prevents LPS-induced liver damage in ethanol-fed animals [29]. These observations reinforce the need to simultaneously target splenic macrophages, a process that is also facilitated by the liposomal delivery system [24,27].
In summary, studies described here provide data on the identification of an siRNA which has high efficacy against rat TNF-α mRNA. The siRNA shows reasonable in vivo efficacy, when administered as a liposomal formulation. With future options to incorporate sequence modifications, it may be possible to enhance the sensitivity and biological half-life of the siRNA. With these options, we believe that liposomal formulations of siRNAs have major potential as therapeutic agents in the treatment of diseases such as alcoholic liver disease, septic shock and rheumatoid arthritis.
Acknowledgments
1. We would like to thank Dr. Yedy Israel, Department of Pharmacological and Toxicological Chemistry, Faculty of Chemical and Pharmaceutical Sciences, and Millennium Institute for Advanced Studies in Cell Biology and Biotechnology, University of Chile, Santiago, Chile, for his valuable comments during the preparation of this manuscript.
2. This work was supported in part by a Grant AA015081 from the National Institute of Alcoholism and Alcohol Abuse.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adachi Y, Bradford BU, Gao W, Bojes H, Thurman RG. Inactivation of Kupffer cells prevents early alcohol-induced liver injury. Hepatology. 1994;20:453–460. [PubMed] [Google Scholar]
- 2.Laskin DL, Gardner CR, Price VF, Jollow DJ. Modulation of macrophage functioning abrogates the acute hepatotoxicity of acetaminophen. Hepatology. 1995;21:1045–1050. [PubMed] [Google Scholar]
- 3.Ishiyama H, Sato M, Matsumara K, Sento M, Ogino K, Hobara T. Proliferation of hepatocytes and attenuation from carbon trtrachloride hepatotoxicity by gadolinium chloride in rats. Pharmacol Toxicol. 1995;77:293–298. doi: 10.1111/j.1600-0773.1995.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 4.Beutler B, Cerami A. Cachectin and tumor necrosis factor as two sides of the same biological coin. Nature. 1986;320:584–588. doi: 10.1038/320584a0. [DOI] [PubMed] [Google Scholar]
- 5.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 6.Decker K. Biologically active products of stimulated liver macrophages (Kupffer cells) Eur J Biochem. 1990;192:245–261. doi: 10.1111/j.1432-1033.1990.tb19222.x. [DOI] [PubMed] [Google Scholar]
- 7.McClain CJ, Cohen DA. Increased tumor necrosis factor production by monocytes in alcoholic hepatitis. Hepatology. 1989;9:349–351. doi: 10.1002/hep.1840090302. [DOI] [PubMed] [Google Scholar]
- 8.Nanji AA, Zhao S, Sadrzadeh SM, Waxman DJ. Use of reverse transcription-polymerase chain reaction to evaluate in vivo cytokine gene expression in rats fed ethanol for long periods. Hepatology. 1994;19:1483–1487. [PubMed] [Google Scholar]
- 9.Kamimura S, Tsukamoto H. Cytokine gene expression by Kupffer cells in experimental alcoholic liver disease. Hepatology. 1995;21:1304–1309. [PubMed] [Google Scholar]
- 10.Elliot MJ, Maini RN, Feldmann M, Kalden JR, Antoni C, Smolen JS, Leeb B, Breeveld FC, Macfarlane JD, Bijl H, Woody JN. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor α (cA2) versus placebo in rheumatoid arthritis. Lancet. 1994;344:1105–1110. doi: 10.1016/s0140-6736(94)90628-9. [DOI] [PubMed] [Google Scholar]
- 11.Miurch SH, Braegger CP, Walker-Smith JA, MacDonald TT. Location of tumor necrosis factor-α by immunohistochemistry in chronic inflammatory bowel disease. Gut. 1993;34:1705–1709. doi: 10.1136/gut.34.12.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michie HR, Manogue KR, Spriggs DR, Revhaug A, O’Dwyer S, Dinerallo CA, Cerami A, Wolf SM, Wilmore DW. Detection of circulating tumor necrosis factor after endotoxin administration. N Eng J Med. 1988;318:1481–1486. doi: 10.1056/NEJM198806093182301. [DOI] [PubMed] [Google Scholar]
- 13.Wanner GA, Muller PE, Ertel W, Bauer M, Menger MD, Messmer K. Differential effect of anti-TNF-α antibody on proinfalmmatory cytokine release by Kupffer cells following liver ischemia and reperfusion. Shock. 1999;11:391–395. [PubMed] [Google Scholar]
- 14.Iimuro Y, Galluchi RM, Luster MI, Kono H, Thurman RG. Antibodies to tumor necrosis factor alpha attenuate hepatic necrosis and inflammation caused by chronic exposure to ethanol in the rat. Hepatology. 1997;26:1530–1537. doi: 10.1002/hep.510260621. [DOI] [PubMed] [Google Scholar]
- 15.Ponnappa BC, Israel Y, Aini M, Zhou F, Russ R, Cao Q, Yiyang Hu, Rubin R. Inhibition of tumor necrosis factor alpha secretion and prevention of liver injury in ethanol-fed rats by antisense oligonucleotides. Biochem Pharmacol. 2005;69:569–577. doi: 10.1016/j.bcp.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Stein CA. Does antisense exist? Nat Med. 1995;1:1119–1121. doi: 10.1038/nm1195-1119. [DOI] [PubMed] [Google Scholar]
- 17.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein E, Denli AM, Hannon GJ. The rest is silence. RNA. 2001;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- 19.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. review. [DOI] [PubMed] [Google Scholar]
- 20.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brummelkamp TJ, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 22.Bertrand JR, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296:1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- 23.Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponnappa BC, Dey I, Tu G, Zhou F, Garver E, Cao Q, Israel Y. In vivo delivery of antisense oligodeoxynucleotides into rat Kupffer cells. J Liposome Res. 1998;8:479–493. [Google Scholar]
- 25.Bautista AP, Schuler A, Spolarics Z, Spitzer JJ. In vivo latex phagocytosis primes the Kupffer cells and hepatic neutrophils to generate superoxide anions. J Leukocyte Biol. 1992;51:39–45. doi: 10.1002/jlb.51.1.39. [DOI] [PubMed] [Google Scholar]
- 26.Szoka F, Papahadjopolos D. Procedure for preparation of liposomes with large internal aqueous space and high capture by reverse-phase evaporation. Proc Natl Acad Sci, (USA) 1978;75:4194–4198. doi: 10.1073/pnas.75.9.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponnappa BC, Dey I, Tu G, Zhou F, Aini M, Cao Q, Israel Y. In vivo delivery of antisense oligonucleotides in pH-sensitive liposomes inhibit lipopolysaccharide-induced production of tumor necrosis factor alpha in rats. J Pharmacol Exp Ther. 2001;297:1129–1136. [PubMed] [Google Scholar]
- 28.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka S, Kumashiro R, Tanikawa K. Role of spleen in endotoxin-induced hepatic injury in chronic alcoholic-fed rats. Liver. 1992;12:306–310. doi: 10.1111/j.1600-0676.1992.tb00578.x. [DOI] [PubMed] [Google Scholar]


