Abstract
Metazoan regeneration is one of the least understood fundamental problems of biology. The lack of progress in understanding this phenomenon at the molecular level has been due to the poor regenerative abilities of the genetic organisms used for developmental studies, as well as the difficulties encountered with molecular and genetic manipulations of the commonly studied vertebrate models (the urodele amphibians). Here, we demonstrate that introduction of double-stranded RNA selectively abrogates gene function in planarians, a classic model of regeneration. The ability to eliminate gene function in a regenerating organism such as the planarian overcomes previous experimental limitations and opens the study of animal regeneration to unprecedented levels of molecular detail.
Among the metazoans, planarians have long been known to possess remarkable regenerative abilities. One example of these abilities was Morgan’s 1898 demonstration that a very small planarian fragment, corresponding to only 1/279th of the intact organism, was capable of regenerating a complete individual (1). Such striking regenerative powers have attracted generations of biologists to the study of this problem, and a vast literature, spanning more than 200 years, exists on the subject (2–6). In addition to their regenerative abilities, flatworms occupy a key position in the evolution of metazoans (7). They are widely acknowledged as being among the simplest organisms possessing three tissue layers (triploblasts), bilateral symmetry, cephalization, and complex organ systems (8). The most commonly studied planarians are the freshwater triclads of the class Turbellaria, phylum Platyhelminthes. Their regenerative properties, combined with their morphological simplicity and phylogenetic position, make planarians ideally suited for the study of regeneration mediated by the formation of a blastema (a bud with well defined epithelial and mesenchymal compartments). Despite their attractiveness as a model system, examination of planarians at the molecular level has just begun (9–11), and their use has been relegated mostly to descriptive, phenomenological studies.
Aiming to develop a model system in which to study the molecular basis of metazoan regeneration and to exploit the remarkable developmental plasticity of planarians, we have begun to screen for genes involved in planarian regeneration (12) and to develop methods with which to study gene function. In lieu of a classical genetic analysis to generate loss-of-function mutants in planarians, we have chosen to test genetic interference caused by double-stranded RNA (RNAi) as recently described in Caenorhabditis elegans (13, 14), Trypanosoma brucei (15), and Drosophila melanogaster (16), as well as in the plants Nicotiana tabaccum and Oryza sativa (17). In these organisms, the introduction of double-stranded (ds) RNA was found to inhibit specifically the expression of the gene from which that RNA was derived. Here, we extend the use of RNAi to eliminate gene expression in planarians, both in regeneration blastemas and in differentiated adult structures.
MATERIALS AND METHODS
Synthesis of dsRNA.
dsRNA was synthesized as described (13). pBluescriptII SK(+) containing the appropriate cDNA inserts was linearized as follows: myosin heavy chain clone M71 (GenBank accession no. AF112359) was digested with HindIII or PstI to generate sense or antisense RNAs with T3 or T7 RNA polymerase, respectively; α-tubulin clone 1 (GenBank accession no. AF112360) was digested with either HindIII or BamHI to synthesize antisense (T3) or sense (T7) RNAs; and opsin clone E42 (GenBank accession no. AF112361) was digested with HindIII or SmaI to synthesize antisense (T3) or sense (T7) RNAs. RNA polymerases were obtained from Promega. After RNA synthesis, the samples were digested with DNase I for 15 minutes at 37°C. The sense and antisense reactions were pooled and annealed for 10 minutes at 37°C, extracted with phenol/chloroform and then chloroform, precipitated with ethanol, and resuspended in 10 μl of diethyl pyrocarbonate-treated H2O. Formation of dsRNA was confirmed by running 0.5 μl of these reactions in a 1.0% agarose gel in TBE (90 mM Tris-borate/2 mM EDTA, pH 8.0).
Microinjection and Immunofluorescence.
Planarian fragments were obtained by cutting animals of ≈6–7 mm with a sterile razor blade at least five times perpendicularly to their anterior/posterior axes. The resulting 1–2 mm fragments were injected with dsRNA or H2O using a Drummond Scientific (Broomall, PA) Nanoject injector and allowed to heal and regenerate at room temperature. To carry out whole-mount immunofluorescence, the fragments were first treated with 2% HCl for 30 seconds at room temperature and then fixed in Carnoy’s fixative for 3 hours. The samples were rinsed briefly in 100% methanol and bleached overnight at room temperature in 6% H2O2 in methanol. The fragments were rehydrated through a graded series of methanol/PBS washes (75%, 50%, 25%) for 5 minutes each, and then blocked in PBS, 0.3% Triton X-100, and 2.5 mg/ml BSA (PBSTB; Sigma) for 5 hours. Anti-planarian myosin heavy chain mAb TMUS-13 and the anti-acetylated tubulin mAb (T-6793; Sigma) were used at 1:10 and 1:1,000 dilutions, respectively, at room temperature for 10–12 hours. At least 4 washes of 2 hours each with PBSTB were carried out before adding the secondary anti-mouse antibody conjugated to Alexa 488 (Molecular Probes) at a 1:400 dilution. After overnight incubation, the samples were washed several hours in PBSTB, mounted in Vectashield (Vector Laboratories) and observed with a Leica TCS NT confocal microscope.
In Situ Hybridization.
Opsin dsRNA-injected samples were processed for whole-mount in situ hybridization as described (10). Digoxigenin-labeled RNA probes were prepared by using the Riboprobe system from Promega and a digoxigenin-labeled ribonucleotide mix from Boehringer Mannheim. Hybridizations were carried out at 56°C overnight. Samples were washed two times for 30 minutes at 56°C in 2× SSC/0.1% Triton X-100, followed by another two 30-minute washes at 56°C in 0.2× SSC/0.1% Triton X-100. An anti-digoxigenin antibody conjugated to alkaline phosphatase (Boehringer Mannheim), and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitroblue tetrazolium (NBT) (GIBCO/BRL) were used to colorimetrically detect probe hybridization. After color development, the samples were post-fixed in 4% paraformaldehyde in PBS, dehydrated in ethanol, infiltrated with LR Gold resin at room temperature, and sectioned with a motorized microtome and glass knife as described by the manufacturer (Polysciences).
RESULTS AND DISCUSSION
dsRNA Disrupts Normal Gene Expression in Regenerating and Terminally Differentiated Planarian Tissues.
To test the efficacy of RNAi in planarians, we chose to investigate its effects on three well characterized tissues: the body-wall musculature (18), the ciliated ventral epithelium (19), and the photoreceptors (20). Of these three, the dynamics of regeneration of the planarian body-wall musculature is perhaps the best characterized to date (21). In the freshwater planarian, the body-wall musculature consists of four subepidermal muscle fiber layers (circular, thin longitudinal, diagonal, and inner longitudinal layers, in that order), and a network of dorsoventral fibers that connects the dorsal and ventral surfaces (21). All of these layers form a highly organized web of myofilaments in the adult organism which, after amputation, regenerates to its original complexity in about 3–5 days at room temperature (Fig. 1 a, c, and e).
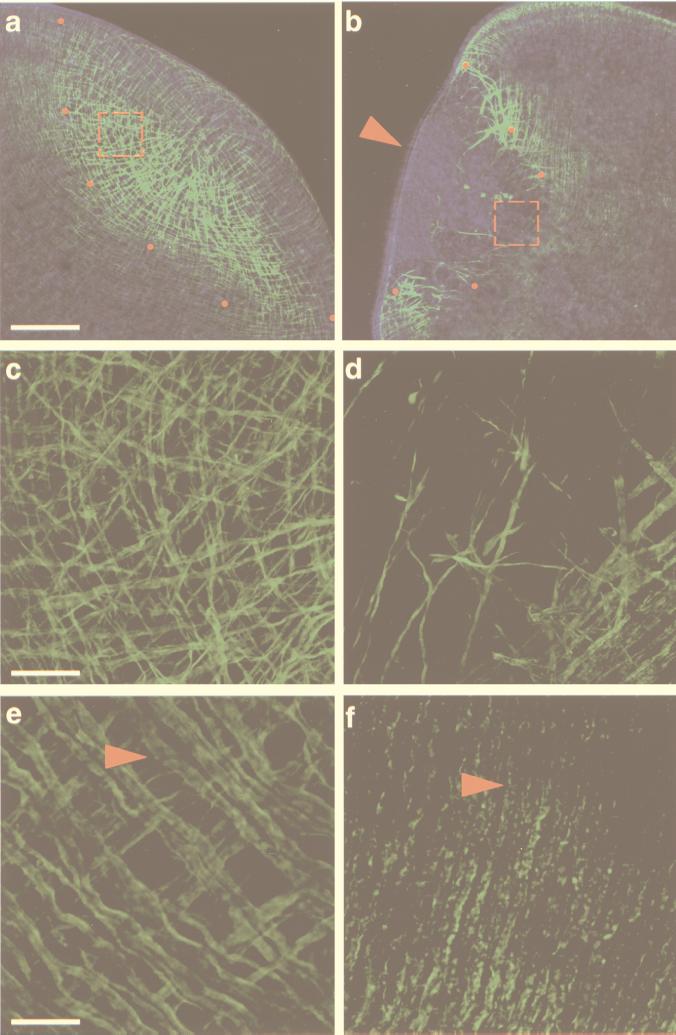
Figure 1.
Effects of myosin dsRNA injection in the planarian S. mediterranea. Confocal projections of body-wall musculature visualized with an anti-myosin heavy chain mAb (21). (a, c, and e) Control animals injected with water. (a and c) The normal dynamics of body-wall muscle regeneration. (e) The intact, terminally differentiated body-wall musculature. (b, d, and f) Myosin dsRNA-injected animals. Note the lack of appropriately regenerated muscle in b. (a and b) Confocal projections of 16 optical sections (1.86-μm intervals). Red dots demarcate the proximal margin of the blastema, and the red, dashed squares denote the areas magnified in c and d. The red arrowhead in b points to the blastemal epithelium imaged by superposing the phase-contrast image on the confocal projection. (c) The regenerating musculature with longitudinal (from left to right), circular (from top to bottom), and diagonal fibers within the blastema. (d) The few disorganized muscle fibers present in the proximal boundary of the blastema are shown with the preexisting body-wall musculature (lower right-hand corner). Note the punctate appearance of these fibers as compared with those shown in c. (c and d) Confocal projections of eight optical sections (0.65-μm intervals). (e and f) Confocal projections of eight optical sections (0.45-μm intervals) of the preexisting body-wall musculature in control and dsRNA-injected animals, respectively. Red arrowheads point to circular fibers. [Bars = 100 μm (a and b), 20 μm (c and d), and 10 μm (e and f).]
We wished to determine whether the introduction of body-wall myosin dsRNA into regenerating planarians could alter the formation of the body-wall musculature. To assay for the effects exerted by the exogenously introduced myosin dsRNA, we used the mAb TMUS-13 (which recognizes planarian myosin heavy chain) to visualize the body-wall musculature (21). dsRNA was synthesized as described (13) by using a 1,186-bp myosin cDNA clone isolated from the diploid, asexual strain of the planarian Schmidtea mediterranea (22). To deliver the myosin dsRNA, individuals of S. mediterranea were cut transversely into 5–6 pieces, and ≈32 nl of a solution containing 109–1011 molecules of myosin dsRNA were injected into the parenchyma and/or the gastrovascular system. The fragments were allowed to heal and regenerate at room temperature for at least 3 days, and were then processed for whole-mount immunofluorescence and analyzed by confocal microscopy (see Materials and Methods).
After 3 days, animals injected with myosin dsRNA show very little, if any, regeneration of the body-wall musculature within their blastemas (54 of 54 with defects) (Fig. 1 b and d), compared with both water-injected (n = 30) and single-stranded RNA-injected (n = 10) controls, in which no defects were observed (Fig. 1 a and c). The few fibers remaining in the blastemas of the myosin dsRNA-injected flatworms likely represent preexisting muscle fibers used to contract the wound surface during the initial stages of wound healing and blastema formation (21). Inspection of the blastema by both Nomarski and phase-contrast microscopy failed to reveal any other gross abnormalities. Superposition of the phase-contrast image on the confocal projection shows the cellular continuity of the epithelium, which completely envelops the muscle-deficient blastema (Fig. 1b, arrowhead).
Examination of the preexisting body-wall musculature also revealed marked defects in 43 of 54 of the injected fragments and a less severe but appreciable deficiency in the remaining injected fragments (11 of 54). In both cases, the phenotype is characterized by the disappearance of TMUS-13 signal from the longitudinal and diagonal fibers as well as by an obvious deterioration of the remaining circular fibers (Fig. 1f), a feature never detected in the water-injected controls (Fig. 1e; arrowheads in Fig. 1 e and f point to circular fibers). These data indicate that the injected dsRNA affects not only the differentiating myogenic lineage in the regeneration blastema, but also the preexisting, terminally differentiated muscle cells in the organism. These results are in agreement with observations in C. elegans, in which both terminally differentiated and embryonic lineages are affected by RNAi (13, 14).
Genetic interference caused by the myosin dsRNA is restricted to the body-wall musculature, and does not appear to affect the musculature of the pharynx (data not shown). This distinction is most likely the result of target specificity of the injected dsRNA. As in C. elegans (23), the musculature of the S. mediterranea pharynx does not express body-wall myosin but rather, a different, unidentified myosin isoform that fails to cross-hybridize to our myosin cDNA probe in whole-mount in situ hybridizations (unpublished results; F. Cebrià, personal communication).
The Effects of dsRNA Injections in Planarians Are Specific.
To confirm the specificity of the observed phenotype, we wanted to determine whether other tissues were being affected by the injected myosin dsRNA. Considering the paucity of molecular markers available for S. mediterranea, we needed to choose a tissue for which antibodies were readily available and whose gross morphology could be scored unambiguously. We chose the ventral epithelium because it is covered with microtubule-containing cilia that can be detected by using a commercially available mAb recognizing acetylated tubulin (24), making disruptions of ciliary morphology easy to score.
First, we examined the effects of myosin dsRNA injections on the ventral ciliated epithelia. Animals were cut, injected, and allowed to heal and regenerate as described above. After injection with myosin dsRNA the musculature was visualized with TMUS-13 and the ventral epithelium with an anti-acetylated tubulin mAb (α-AcTub). Fig. 2a shows a regeneration blastema 7.5 days postinjection. Even after 7.5 days, the musculature phenotype observed at earlier time points persists, and only a few, very disorganized muscle fibers can be detected within the blastema (Fig. 2a, green). However, the differentiation of the blastemal epithelium proceeded unaffected, as shown by the presence of cilia at the edge of the blastema (Fig. 2a, red, arrowhead). Notably, the ventral epithelium also regenerated normally in all analyzed muscle-deficient blastemas (Fig. 2b; n = 12), and was indistinguishable from epithelium in control injections (Fig. 2e; n = 24).
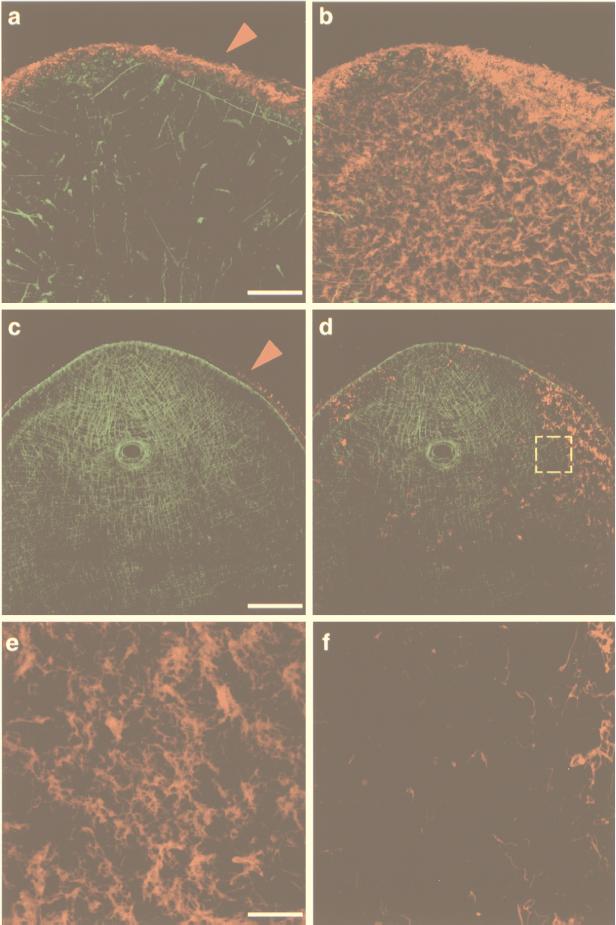
Figure 2.
Myosin and α-tubulin dsRNA injections in S. mediterranea. Confocal projections of body-wall musculature and ventral ciliated epithelium visualized with anti-myosin heavy chain and anti-acetylated tubulin (α-AcTub) mouse mAbs, respectively. The tubulin signal was pseudo-colored in red and was separated from the musculature signal by projecting only optical sections corresponding to the blastema’s ventral surface. (a) Projection of the complete subepithelial musculature (in green) in a 7.5-day cephalic blastema of an animal injected with myosin dsRNA (16 optical sections, 1.2-μm intervals). Note the disorganization of the musculature. The epithelium surrounding each of the optical sections and recognized by the α-AcTub antibody was pseudo-colored in red (arrowhead) and appears normal. (b) Superposition of the projected ventral epithelium (eight optical sections, 0.48-μm intervals) on the confocal projection shown in a. (c) Projection of the complete subepithelial musculature (in green) in a caudal blastema of an animal injected with α-tubulin dsRNA (16 optical sections, 1.41-μm intervals). No muscle defects are observed. The circular aperture is the normally regenerated pharyngeal opening. As in a, the epithelium recognized by the α-AcTub antibody was pseudo-colored in red (arrowhead). (d) Superposition of the projected ventral epithelium (eight optical sections, 0.35-μm intervals) on the confocal projection shown in c showing the disruption of the ciliated epithelium. (e) Confocal projection of 10 optical sections (0.21-μm intervals) of a normal (water-injected) caudal blastema. (f) The area demarcated by the yellow, dashed square in d shown at higher magnification. [Bars = 50 μm (a and b), 100 μm (c and d), and 20 μm (e and f).]
Second, to eliminate the possibility that the effect on the musculature was nonspecific and would be caused by the injection of any dsRNA, we injected α-tubulin dsRNA into planarian pieces as described above (n = 24) in amounts ranging from 108 to 1010 molecules. The injected fragments were allowed to heal and to regenerate for 3–7.5 days, processed for whole-mount immunofluorescence, and scored for immunoreactivity with TMUS-13 and α-AcTub antibodies using confocal microscopy. Fig. 2c shows a 7.5-day blastema injected with α-tubulin dsRNA in which a complete, normally regenerated body-wall musculature is present (in green). Thus, injected α-tubulin dsRNA did not interfere with body-wall muscle regeneration.
In contrast, the edge epithelium contained abnormally low levels of α-tubulin (compare arrowheads in Fig. 2 a and c), and the ventral epithelium displayed a dramatic reduction in the number of cilia surrounding the blastemas and very few, if any, cilia in the regenerated ventral surface of the same blastemas (Fig. 2 d and f, red). Higher magnification views of the ventral epithelium revealed that in the α-tubulin-injected fragments, clusters of cilia were sparse, and an abnormal frequency of what appear to be single cilia were present (Fig. 2f) when compared with the water-injected controls (Fig. 2e) and the myosin dsRNA-injected fragments (Fig. 2b). The data presented in Fig. 2 clearly demonstrate the efficiency and specificity of injected dsRNA as a means of inhibiting gene expression during planarian regeneration.
dsRNA Acts by Reducing the Normal Accumulation of Endogenous RNA Transcripts.
To ascertain whether dsRNA interference acts by decreasing or eliminating endogenous mRNA transcripts, as reported for C. elegans (13, 25) and T. brucei (15), we examined the effect of dsRNA on the endogenous expression levels of opsin mRNA. This molecule is expressed exclusively in the ≈25 retinular cells present per planarian photoreceptor (Fig. 3 a, purple and b, blue). S. mediterranea head fragments with intact photoreceptors were injected with a 447-bp opsin dsRNA synthesized from an S. mediterranea cDNA clone. Approximately 108–109 copies of this molecule were injected per head. Animals were fixed and processed for in situ hybridization (10) at several time points after injection. Representative data are shown in Fig. 3.
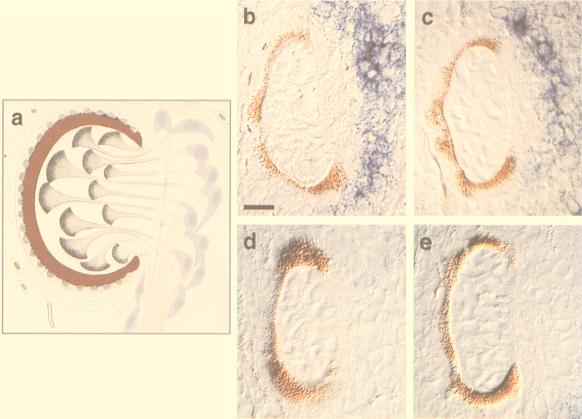
Figure 3.
Effect of opsin dsRNA injection on the levels of opsin mRNA in the retinular cells of S. mediterranea photoreceptors. (a) The components of the planarian photoreceptor (modified from von Graff) (26). The pigment cup is colored in red and the cell bodies of the light-sensing retinular cells in purple; their rhabdomeres project into the pigment cup. (b) Opsin levels detected 24 hours after injecting heads with water; the localization of opsin mRNA is revealed by the blue signal. (c–e) Opsin levels detected 6, 12, and 24 hours, respectively, after injecting the head fragments with opsin dsRNA. (b–e) Nomarski images of 4-μm plastic sections through injected head fragments processed for whole-mount in situ hybridization (10) to detect opsin transcripts. (Bar = 10 μm.)
A slight decrease in opsin mRNA levels in retinular cells was seen 6 hours postinjection (Fig. 3c, n = 4), and by 12 hours, opsin mRNA was not detected in the retinular cells in 8 of 11 injected heads (Fig. 3d). By 24 hours postinjection, opsin mRNA was completely absent from the retinular cells in 12 of 12 injected heads (Fig. 3e). By contrast, the expression levels of opsin in the photoreceptors of the water-injected control heads (n = 9) remained unchanged throughout the course of the experiment (Fig. 3b). In addition, no modulation of the opsin transcript level was observed when α-tubulin dsRNA was injected at equal (108–109 molecules) or higher (109–1011 molecules) amounts (data not shown).
As observed in C. elegans, D. melanogaster, and T. brucei and the plants N. tabaccum and O. sativa, introduction of dsRNA in S. mediterranea is capable of specifically repressing gene expression. Although the precise mechanism by which dsRNA inhibits gene expression is unclear, it appears to result in the degradation of the corresponding endogenous mRNA before its translation (15, 25). In C. elegans, the site of dsRNA injection does not necessarily correspond to the site of action of the dsRNA: injection into the gut produces germ-line defects, and injection into the gonad produces somatic defects (13). Likewise, in planarians, injections into either the gut or the parenchyma produce defects elsewhere in the body. These results suggest that in both of these organisms, a transport mechanism must be present to deliver the dsRNA from extra- to intracellular compartments.
Our present results provide strong evidence for the effectiveness and potency of RNAi in the planarian, a model organism for studying metazoan regeneration that has been inaccessible to genetic manipulations. The feasibility of carrying out loss-of-function assays in S. mediterranea signifies that a functional characterization of key molecules controlling regeneration in metazoans is now possible. Consequently, the study of the simple, triploblastic planarian, combined with a molecular analysis of its regeneration-modulated genes, will shed light on one of the oldest unresolved problems of biology and may ultimately help us identify the molecular factors that make regeneration permissive in some, but not in all, animals.
Acknowledgments
We thank Dr. Rafael Romero Benedí for the TMUS-13 antibodies, Francesc Cebrià and John Henry Sánchez for cloning and sequencing the S. mediterranea myosin and opsin clones used for these studies, and Mike Sepanski for producing the plastic sections of planarian photoreceptors. We also thank our colleagues at the Carnegie Institution, Drs. Donald D. Brown and Joseph G. Gall for their critical comments on this manuscript and Dr. Andy Fire for suggesting the use of RNAi in planarians. We extend our gratitude to Dr. Jaume Baguñà for sharing with us his extensive knowledge of planarian biology. Comments provided by Dr. Tatjana Piotrowski during the final editing process are also kindly acknowledged. This work was funded by National Institutes of Health National Institute of General Medical Sciences RO1 GM57260-01 to A.S.A and by postdoctoral fellowships from the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (DRG-1322), and National Institutes of Health (1F32-GM19539-01) to P.A.N.
ABBREVIATIONS
- RNAi
RNA-mediated genetic interference
- dsRNA
double-stranded RNA
Footnotes
References
- 1.Morgan T H. Arch Entw Mech Org. 1898;7:364–397. [Google Scholar]
- 2.Morgan T H. Regeneration. New York: Macmillan; 1901. [Google Scholar]
- 3.Child C M. Patterns and Problems of Development. Chicago: Univ. of Chicago Press; 1941. [Google Scholar]
- 4.Brøndsted H V. Planarian Regeneration. London: Pergamon; 1969. [Google Scholar]
- 5.Baguñà J, Romero R, Saló E, Collet J, Auladell C, Ribas M, Riutort M, Garcia-Fernàndez J, Burgaya F, Bueno D. In: Experimental Embryology in Aquatic Plants and Animals. Marthy H-J, editor. New York: Plenum; 1990. pp. 129–162. [Google Scholar]
- 6.Baguñà J. In: Cellular and Molecular Basis of Regeneration: From Invertebrates to Humans. Ferretti P, Géraudie J, editors. New York: Wiley; 1998. pp. 135–165. [Google Scholar]
- 7.Willmer P. Invertebrate Relationships. New York: Cambridge Univ. Press; 1994. [Google Scholar]
- 8.Brusca R C, Brusca G J. Invertebrates. Sunderland, MA: Sinauer; 1990. [Google Scholar]
- 9.Garcia-Fernàndez J, Baguñà J, Saló E. Development. 1993;118:241–253. doi: 10.1242/dev.118.1.241. [DOI] [PubMed] [Google Scholar]
- 10.Umesono Y, Watanabe K, Agata K. Dev Growth Differ. 1997;39:723–727. doi: 10.1046/j.1440-169x.1997.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- 11.Agata K, Soejima Y, Kato K, Kobayashi C, Umesono Y, Watanabe K. Zool Sci. 1998;15:433–440. doi: 10.2108/zsj.15.433. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez Alvarado A, Newmark P A. Wound Repair Regen. 1998;6:413–420. doi: 10.1046/j.1524-475x.1998.60418.x. [DOI] [PubMed] [Google Scholar]
- 13.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Tabara H, Grishok A, Mello C C. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- 15.Ngo H, Tschudi C, Gull K, Ullu E. Proc Natl Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennerdell J R, Carthew R W. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 17.Waterhouse P M, Graham M W, Wang M-B. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen K J. Wilhelm Roux Arch Entw Org. 1972;169:134–169. doi: 10.1007/BF00649889. [DOI] [PubMed] [Google Scholar]
- 19.Morita M, Best J B. J Exp Zool. 1974;187:345–373. doi: 10.1002/jez.1401870305. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter K, Morita M, Best J. Cell Tissue Res. 1974;148:143–158. doi: 10.1007/BF00224579. [DOI] [PubMed] [Google Scholar]
- 21.Cebrià F, Vispo M, Newmark P, Bueno D, Romero R. Dev Genes Evol. 1997;207:306–316. doi: 10.1007/s004270050118. [DOI] [PubMed] [Google Scholar]
- 22.Benazzi M, Baguñà J, Ballester R, Puccinelli I, Del Papa R. Boll Zool. 1975;42:81–89. [Google Scholar]
- 23.Fire A, Albertson D, Harrison S W, Moerman D G. Development. 1991;113:503–514. doi: 10.1242/dev.113.2.503. [DOI] [PubMed] [Google Scholar]
- 24.LeDizet M, Piperno G. Methods Enzymol. 1991;196:264–274. doi: 10.1016/0076-6879(91)96025-m. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery M K, Xu S, Fire A. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Graff L, editor. Vermes. Ic: Turbellaria. Vol. 4. Leipzig, Germany: C. F. Winter’sche Verlangshandlung; 1915. [Google Scholar]