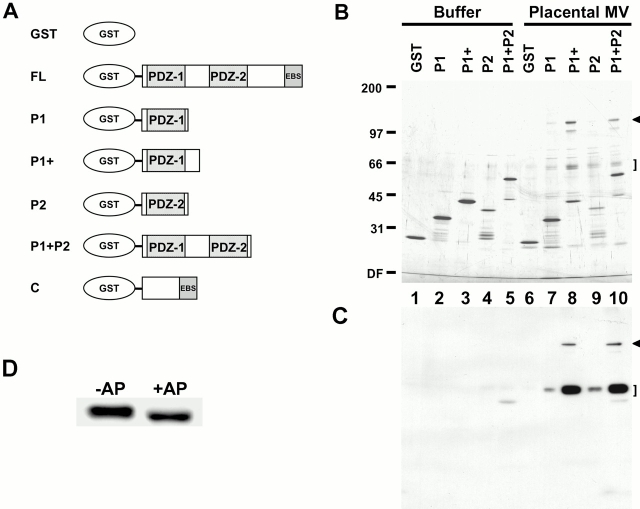
Figure 1.
Identification of EBP50 PDZ domain–binding candidates. (A) Summary of GST–EBP50 fusion proteins used for affinity chromatography. The cDNA sequences encoding human EBP50 residues 1–358 (FL), residues 1–97 (P1), residues 1–138 (P1+), residues 138–248 (P2), residues 1–248 (P1 + P2), and residues 241–358 (C) were expressed as soluble recombinant proteins fused to GST. These proteins were purified, immobilized on glutathione–agarose beads, and then used for affinity chromatography. (B) Affinity chromatography. Buffer or total detergent soluble extracts of placental microvilli were mixed with GST–agarose beads or GST–EBP50–agarose beads containing the first PDZ domain (P1 or P1+), the second PDZ domain (P2), or both in tandem (P1 + P2). The beads were then washed extensively in buffer at 0.3 M NaCl, and bound proteins were eluted and resolved on a 6–20% silver-stained gradient SDS gel. (C) One-fourth the amount of the samples shown in B were resolved by SDS-PAGE, transferred to PVDF, and then probed with biotinylated EBP50. Arrowheads and brackets indicate specific 120-kD and 64/65-kD binding candidates, respectively. The mobilities of molecular mass standards in kD are indicated at left. DF, dye front. (D) The heterogeneity of the 64/65-kD candidate is due to phosphorylation. A PDZ domain affinity bead eluate from B that was enriched in the 64/65-kD binding candidate was incubated in the presence (+AP) or absence (−AP) of calf intestinal alkaline phosphatase and then resolved by SDS-PAGE on a 10% gel, transferred to PVDF, and overlaid with biotinyl EBP50 probe as in C.