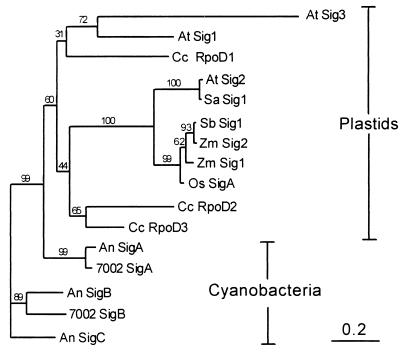
Figure 2.
Phylogenetic analysis of five σ factors from cyanobacteria, three from red algae, four from monocotyledonous plants, and four from dicotyledonous plants. All 16 sequences were aligned by using the pileup program of GCG package. Only the highly conserved 110 amino acid residues (containing subdomains 2.1–2.4 as shown in Fig. 1) were chosen for the calculation of the evolutionary distances by the protdist program of the phylip package by using the pam matrix method. The unrooted tree was constructed from the distance data by the neighbor program in the same package by using the neighbor-joining method. Bootstrap analysis was performed by resampling the data set 100 times, and the number at each node of the tree indicates the supporting percentage of the branching pattern. The scale bar represents 20% substitution, which is proportional to horizontal distance of the tree branches. The higher plant sequences are the same as in Fig 1. Cc RpoD1, Cc RpoD2 and Cc RpoD3, σ factors of the red alga C. caldarium (L42639; AF050634; D83179). An SigA, An SigB, and An SigC, σ factors of Anabaena sp. PCC 7120 (M60046; M95760; M95759) (25, 41). 7002 SigA and 7002 SigB, σ factors of Synechococcus sp. PCC 7002 (U15574; U82435) (21, 42).