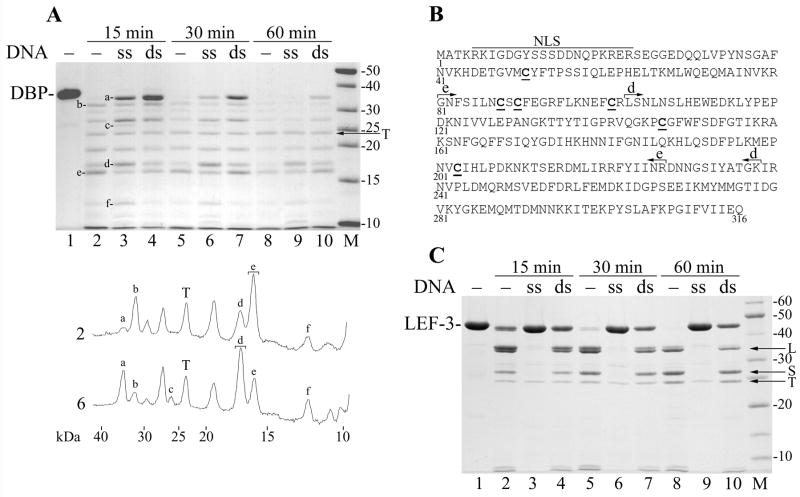
Figure 6.
Protection of DBP and LEF-3 against trypsin by ssDNA and dsDNA. (A) Three 35-μl mixtures containing 3.6 μM DBP, 50 mM Tris-HCl, pH 7.5, 0.1 M NaCl, 12.5% glycerol, and 5 mM DTT were assembled on ice. The mixtures #1, #2, and #3 contained no DNA, 100 μg/ml of M13 ssDNA, and 100 μg/ml of M13 dsDNA, respectively. The mixtures were incubated for 15 min at 25ºC to form complexes of DBP with DNA. Then trypsin (12 μg/ml) was added and the incubation was continued. 10-μl portions were removed from the mixtures at 15 min (lanes 2–4), 30 min (lanes 5–7), and 60 min (lanes 8–10). The reactions were terminated by the addition of 6.1 μl of loading buffer (3 mM PMSF, 0.65 M NaCl, 190 mM Tris, pH 6.8, 4.3 M 2-mercaptoethanol, 6% SDS) followed by boiling for 3 min, and then analyzed by SDS-13% PAGE. Trypsin was not added to the time zero reaction (lane 1). Densitometry of lanes 2 and 6 is shown below the panel. Symbols a to f correspond to respective bands in the gel. Brackets show the bands taken for MALDI/TOF analysis. (B) Location of major tryptic fragments in the DBP sequence. The putative bipartite nuclear localization sequence (NLS, residues 5–23), fragments e (residues 103–238) and d (residues 81–227) are shown. The Cys residues are underlined. (C) Three 35-μl mixtures containing 4.5 μM LEF-3 and other ingredients as in the experiment shown in panel A were assembled on ice. The mixtures #1, #2, and #3 contained no DNA, 40 μg/ml of M13 ssDNA, and 40 μg/ml of M13 dsDNA, respectively. After pre-incubation of LEF-3 with DNA, trypsin (10 μg/ml) was added and the portions were removed from the mixtures at 15 min (lanes 2–4), 30 min (lanes 5–7), and 60 min (lanes 8–10) and processed as described above. Trypsin was not added to the time zero reaction (lane 1). The arrows show position of trypsin (T), and fragments L and S of LEF-3.