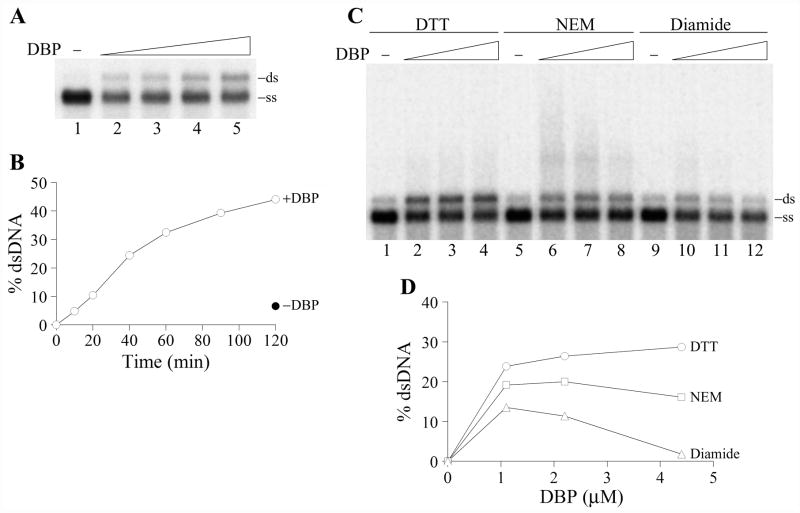
Figure 8.
Renaturation activity of DBP. (A) The renaturation assay was carried out in 10-μl reaction mixtures containing 2 nM 32P-labeled 2289-bp fragment of RF DNA M13mp9, 20 mM Tris-HCl, pH 7.5, 75 mM NaCl, 3 mM CaCl2, 10% glycerol, 100 μg/ml BSA, and 2 mM DTT. DNA was denatured by boiling for 5 min followed by chilling on ice before the addition to the mixtures. DBP was added in the following concentrations: control, no DBP (lane 1), 0.55 μM (lane 2), 1.1 μM (lane 3), 2.2 μM (lane 4), and 4.4 μM (lane 5). The reactions were incubated for 1 h at 37ºC, then EDTA (12 mM) and SDS (0.5%) were added, and the samples were treated with proteinase K (0.5 mg/ml) for 15 min at 22ºC and analyzed by electrophoresis in a 1% agarose gel. The position of double-stranded DNA (ds) and single-stranded DNA (ss) in the gel is shown on the right. (B) Time course of the renaturation reaction promoted by DBP at 37ºC. Reactions containing 2.2 μM DBP were assembled and analyzed as in panel A. The filled symbol shows the amount of dsDNA in the reaction mixture incubated for 120 min at 37ºC in the absence of DBP. (C) and (D) Redox sensitivity of the renaturation reaction promoted by DBP. The renaturation assay was carried out as in panel A. The reactions contained the following redox agents, 5 mM DTT (lanes 1–4), 10 mM N-ethylmaleimide (NEM) (lanes 5–8), and 10 mM diamide (lanes 9–12). DBP was added in the following concentrations: control, no DBP (lanes 1, 5, and 9), 1.1 μM (lanes 2, 6, and 10), 2.2 μM (lanes 3, 7, and 11), and 4.4 μM (lanes 4, 8, and 12). Panel D shows quantification of the data shown in panel C. The amounts of dsDNA generated in the absence of DBP were taken as zero points.