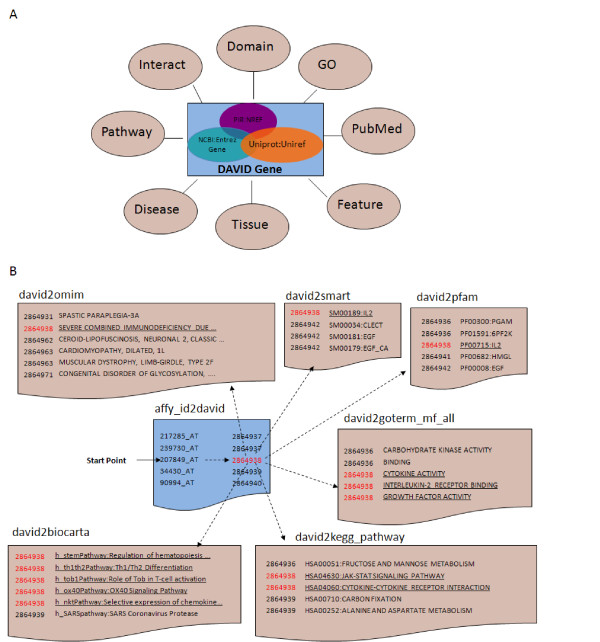
Figure 2.
The gene-centric DAVID Knowledgebase in a simple pair-wise text format centralized by DAVID gene identifiers. A. A hypothetical graph shows that a wide-range of annotation categories are collected and integrated by the DAVID gene cluster IDs. B. A real example demonstrates how the pair-wise text data structures are used in the DAVID Knowledgebase. Each independent annotation source or gene identifier system is separated into independent files in the same pair-wise format of "DID-to-annotation". In this example, a user starts with an Affymetrix identifier (affy_id) 207849_at (IL2). The first step is to obtain the corresponding DAVID gene identifier (DID:2864938). Then, with this DID (red), the annotation terms of interest (underlined) in different source files (OMIM, SMART, Pfam, GO Molecular Function, KEGG Pathway, BioCart Pathway, etc.) can be queried sequentially.