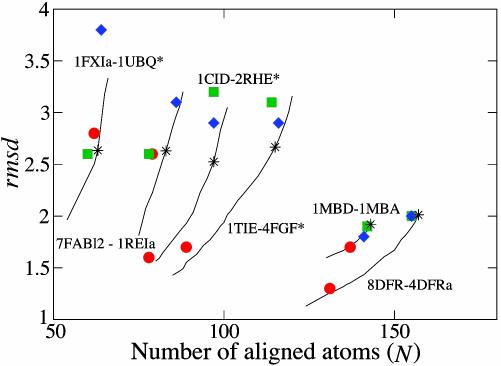
Fig. 3.
Alignment results for a set of protein pairs in terms of rmsd and number of aligned atoms (N). The results from Yale (red circles), Dali (green squares), CE (blue diamonds), and our method (black asterisks) are plotted. For comparison, different rmsd–N pairs for our algorithm, obtained by varying the size of the gap penalty parameter, are shown (solid lines). Our method gives the largest improvements compared to the others for protein pairs from the 10 difficult structures (protein pairs marked with asterisks).