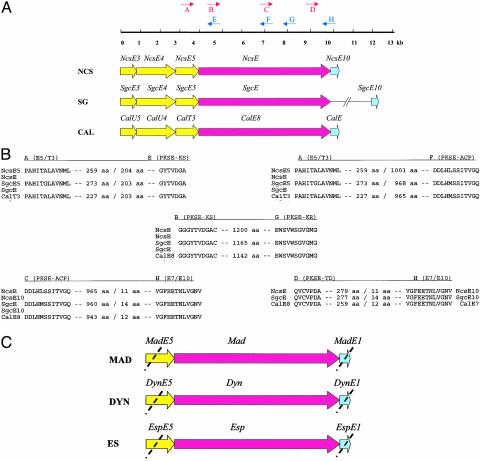
Fig. 2.
(A) Schematic representation of primer design for PCR-based amplification of enediyne PKS (PKSE) fragments. The PKSE and associated genes shown constitute the minimal PKSE cassettes from the 1, 2, and 6 biosynthetic loci, respectively, which are conserved among all enediyne gene clusters known to date. (B) The amino acid motifs used for the design of the degenerate primers A–H, the DNA sequences of which are illustrated in Table 1. The numbers between the motifs indicate the distance in amino acids. The noncoding regions between ORFs, represented by a slash, are <10 nt. (C) Amplified products using the PCR-based approach from genomic DNA isolated from the 3-, 7-, and 8-producing organisms to give madE, dynE, and espE, respectively.