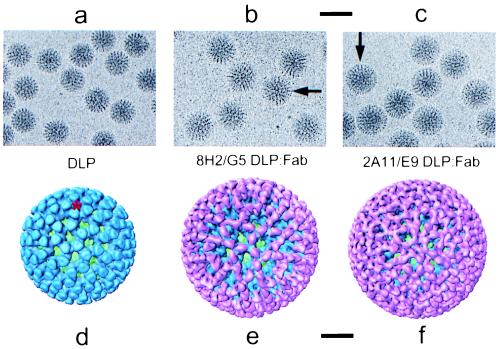
Figure 2.
(a–c) Electron cryomicrograph images of native DLPs, 8H2/G5 DLP/Fab complexes, and 2A11/E9 DLP/Fab complexes. These images were recorded at a defocus of −2.2 μm. (Scale bar, 1,000 Å.) The binding of the Fabs form what appears to be an additional capsid layer on the DLP, visible in the images as a ring of mass density extending outward from a radius of 350 Å (arrows in b and c). (d–f) Surface representations of the 3D structures of the native DLP, 8H2/G5 DLP/Fab complex, and 2A11/E9 DLP/Fab complex, viewed along the icosahedral 3-fold axis. The inner capsid protein VP2 is shown in green, outer capsid protein VP6 in blue, and bound Fabs in pink. This color scheme is used in all figures. In the native DLP, the 780 molecules of VP6 are assembled as trimers organized on a T = 13 (levo) icosahedral lattice (9). One of the channels through which mRNA is released during transcription (3) is indicated with a red star. These channels lie along the icosahedral 5-fold axes. (Scale bar, 200 Å.)