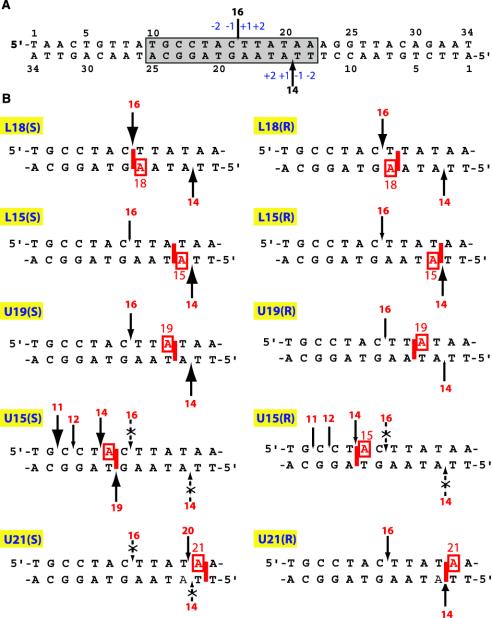
Fig. 2.
(A) Sequence of the oligonucleotide substrate derived from the simian virus 40 nuclear matrix-associated region. The Top2 cleavage site stabilized by VP-16 at position 4265 (34) generates a 16-mer oligonucleotide when the upper strand (U) is 5′-end-labeled and a 14-mer when the lower strand (L) is 5′-end-labeled. The shaded box corresponds to the sequences represented in B. The sizes of the arrows correspond to Top2 cleavage in the absence of VP-16. (B) Summary of the position-specific effects of intercalated BaP DE-2 dA adducts (only the boxed sequence in A is shown). The sizes of the arrows are in proportion to the intensity of Top2-mediated cleavage observed in the presence of the adduct independent of VP-16. Dotted arrows with x symbols denote suppression of Top2-mediated DNA cleavage regardless of the presence or absence of VP-16.