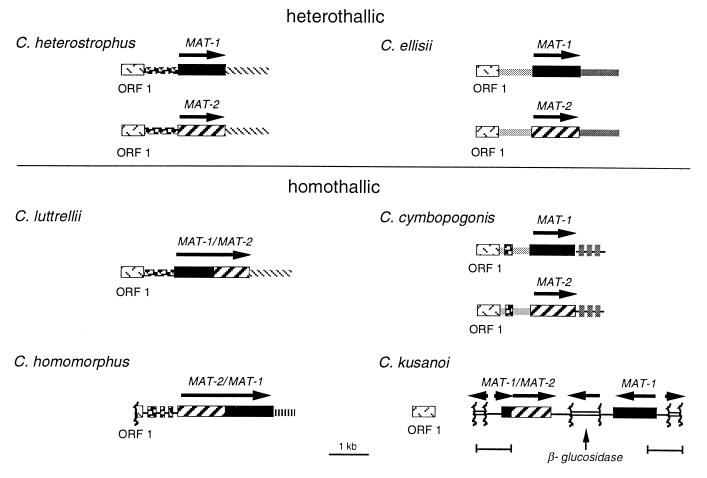
Figure 2.
Organization of MAT in heterothallic and homothallic species. The arrangement is identical in all heterothallic species examined to date, including C. heterostrophus and C. ellisii (shown here), C. carbonum, C. victoriae, C. intermedius, and asexual B. sacchari and A. alternata (not shown, see Fig. 5). Organization of each homothallic locus is unique, as described in the text. Textures of boxes are as in Fig. 1 for MAT-1, MAT-2, and ORF1; gene encoding β-glucosidase (open boxes); all other textures represent noncoding sequences 5′ or 3′ of MAT that are either unique to a particular species or common to more than one. Arrows indicate direction of transcription. Tightly linked to MAT in all species (except C. kusanoi) is a highly conserved ORF (ORF1) that shows similarity to a Saccharomyces cerevisiae ORF (GenBank accession no. U22383) of unknown function. Note that all genes are linked or fused, except that linkage has not yet been detected between the C. cymbopogonis MAT genes (which reside in the same nucleus unlike the heterothallic ones which reside in separate nuclei) or between ORF1 and C. kusanoi MAT.