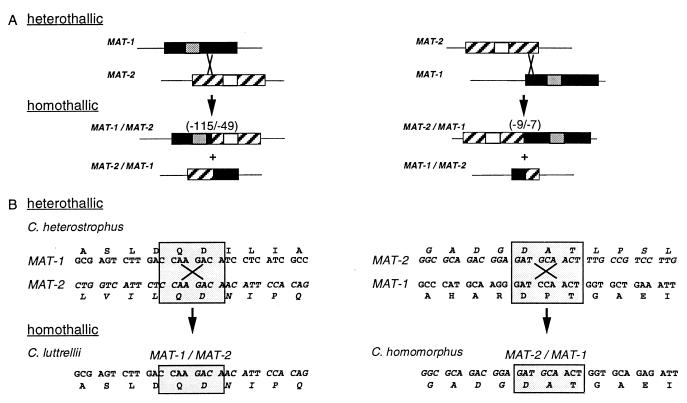
Figure 3.
Models for evolution, by recombination events, of fused homothallic MAT genes in C. luttrellii and C. homomorphus from opposite heterothallic MAT genes in C. heterostrophus (the heterothallic progenitor of C. homomorphus is unknown). (A) Misalignment of homologous flanking sequences could bring into register short islands of identity between the largely dissimilar MAT idiomorphs. A homologous recombination event at the point of identity would result in two fused MAT genes, both incomplete with respect to their heterothallic counterparts. If the crossover point were on either side of the DNA binding region (14), one fusion product would have both DNA-binding motifs and one would have neither. The number of amino acids eliminated (C. luttrellii, 115 from the 3′ end of MAT-1, 49 from the 5′ end of MAT-2; C. homomorphus, 9 from the 3′ end of MAT-2, 7 from the 5′ end of MAT-1) depends on the position of the crossover point (compare A Left with A Right). Textures are described in Fig. 1; small boxes within idiomorphs represent DNA-binding motifs; gray, α-box in MAT-1, white, HMG box in MAT-2 (14). (B) Inspection of the actual nucleotide sequences of the C. heterostrophus MAT-1 and MAT-2 genes reveals an 8-bp region of complete identity (shaded box, B Left) and a 9-bp region, with one mismatch (shaded box, B Right), corresponding to the C. luttrellii and C. homomorphus fusion points, respectively. Recombination in the Upper Left box creates precisely the sequence found in the C. luttrellii MAT-1/MAT-2 fused gene (Lower Left box); the left side of the fused sequence is similar to C. heterostrophus MAT-1 and the right side to MAT-2. Recombination in the Upper Right box creates the C. homomorphus MAT-2/MAT-1 fused gene (Lower Right box). Single letters above or below codons are standard amino acid abbreviations.