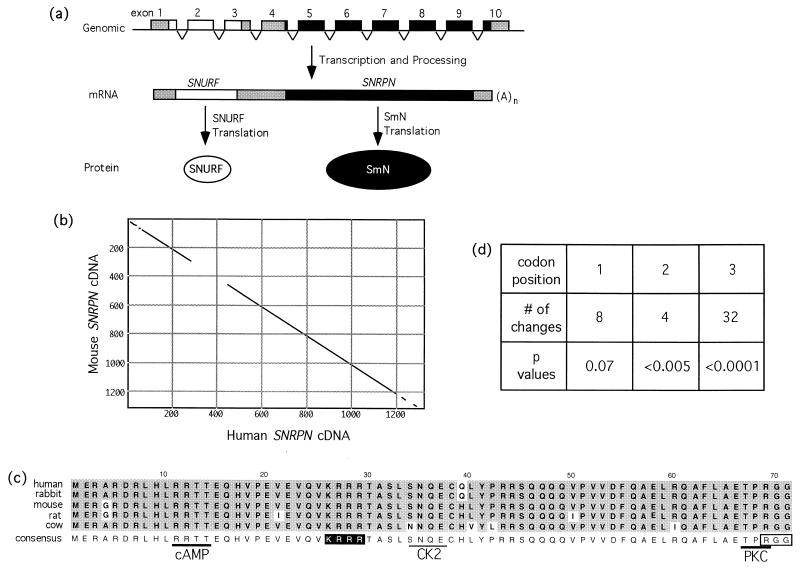
Figure 1.
SNURF–SNRPN gene structure and sequence analysis. (a) Schematic representation of the bicistronic SNURF–SNRPN locus depicting the progression from a contiguous nonoverlapping series of exons to a single transcript with two ORFs, to independent protein products. Open and filled structures correspond to SNURF or SNRPN, respectively, whereas the shaded regions indicate untranslated sequences. (b) Dot-plot analysis of full-length mouse and human SNRPN cDNAs; conserved sequences (window = 30, 65% minimum) appear as a diagonal line. (c) Amino acid alignment of putative SNURF proteins from five eutherian mammals. The derived consensus is shown below the compilation; residues consistent with the consensus are shaded in gray, changes are unshaded. Potential features appended to the consensus are: a nuclear localization signal (white letters on a black background), a C-terminal RGG motif (boxed), and phosphorylation sites that are absolutely conserved (heavy underline; cAMP, cAMP-dependent kinase; PKC, protein kinase C) or partially conserved (light underline; CK2, casein kinase II). (d) Codon position of the nucleotide changes that occur in the SNURF coding sequence of the five species examined. P values were derived from a χ2 analysis at each position.