Abstract
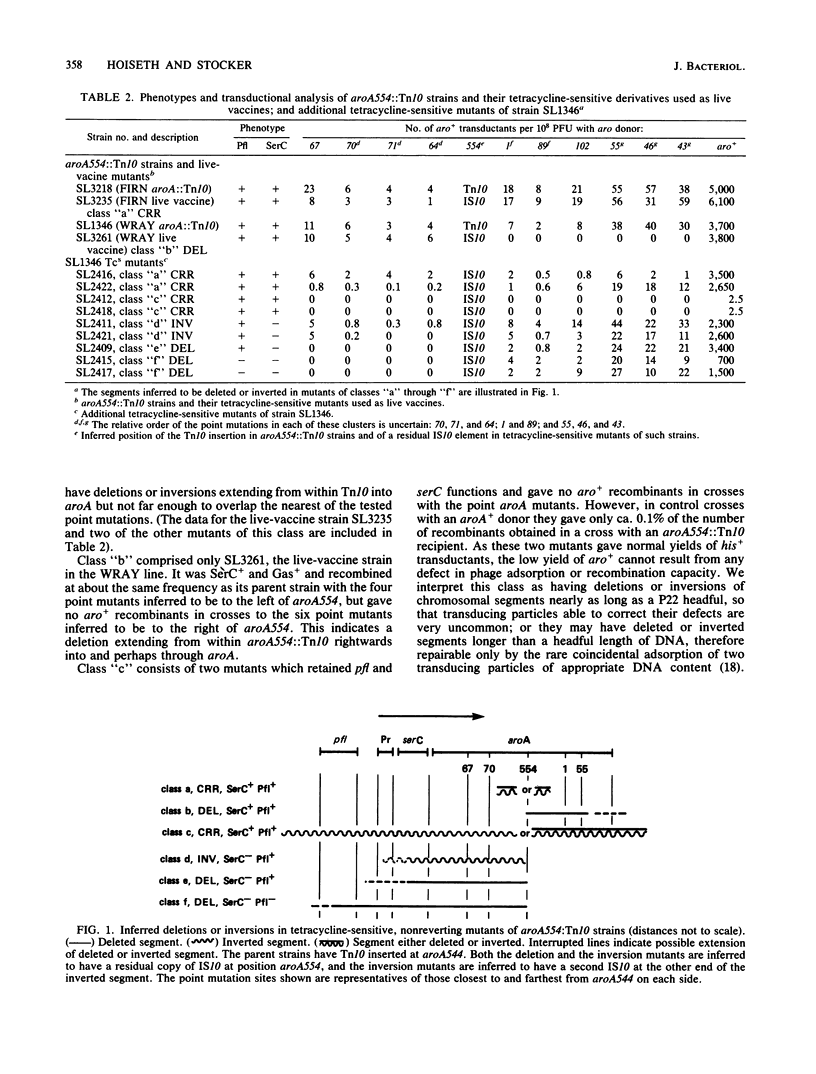
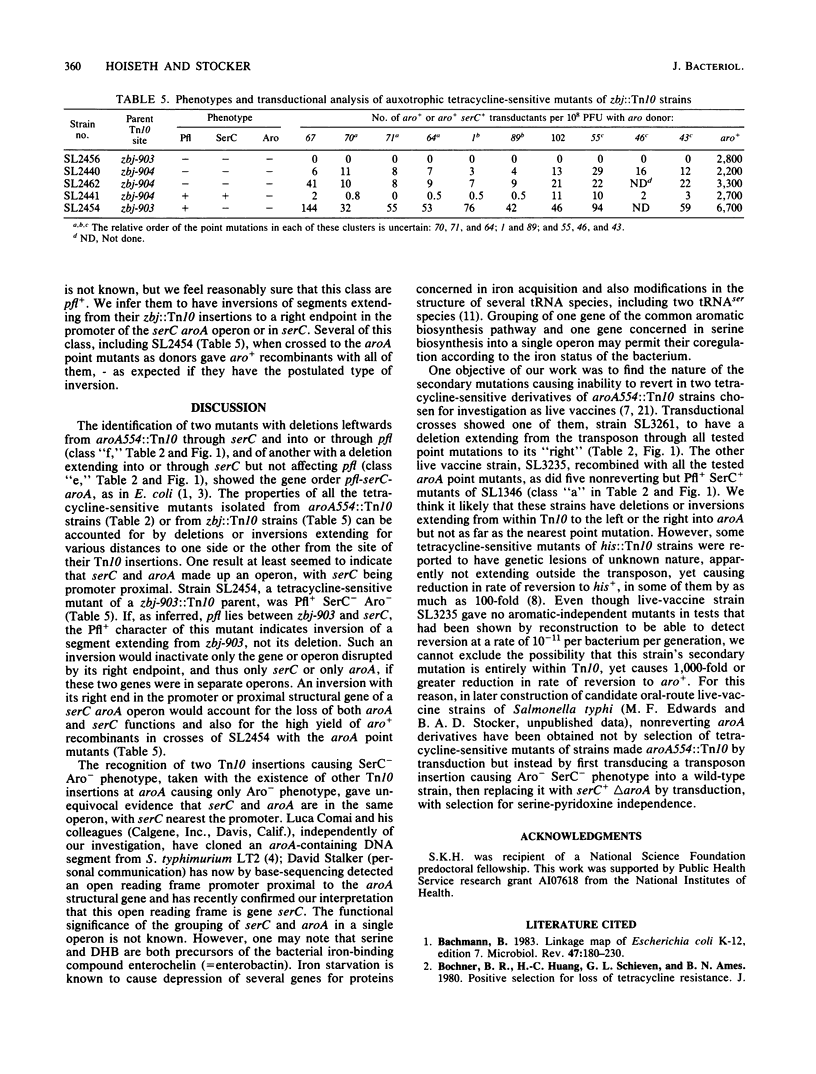
Genetic analysis of aroA554::Tn10 derivatives of two mouse-virulent Salmonella typhimurium strains, "FIRN" and "WRAY," and of a nonreverting derivative of each constructed for use as a live vaccine, showed the site of the insertion among mapped aroA point mutants. The WRAY live-vaccine strain gave no aro+ recombinants in crosses with aroA point mutations to one side of the insertion, indicating a deletion from Tn10 through the sites of these point mutations. The FIRN live-vaccine strain gave wild-type recombinants with all tested point mutants; it probably has a deletion or inversion extending from Tn10 into aroA but not as far as the nearest point mutation. Some tetracycline-sensitive mutants of aroA554::Tn10 strains required serine and pyridoxine, indicating loss of serC function, and some that were found to be SerC- did not produce gas from glucose, indicating a loss of pfl function. These results show the gene order pfl-serC-aroA, as in Escherichia coli. Ampicillin enrichment applied to pools of tetracycline-sensitive mutants of strains with Tn10 insertions near aroA (i.e., zbj::Tn10 strains) yielded Aro- SerC- Pfl-, Aro- SerC+ Pfl+, and Aro- SerC- Pfl+ mutants but none which were Aro+ SerC-. All of the mutants are explicable by deletions or inversions extending clockwise from zbj::Tn10 into or through an operon comprising serC (promoter-proximal) and aroA. Such an operon was also shown by the identification of two Tn10 insertions causing phenotype Aro- SerC-, each able to revert to Aro+ SerC+ by precise excision. serC corresponds to the open reading frame promoter-proximal to aroA that was identified elsewhere by base sequencing of a cloned aroA segment of S. typhimurium (Comai et al., Science 221:370-371, 1983). Both serine and chorismate are precursors of enterochelin; this may be why serC and aroA are in a single operon.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. J., Low B., Konigsberg W. H. Close linkage of the genes serC (for phosphohydroxy pyruvate transaminase) and serS (for seryl-transfer ribonucleic acid synthetase) in Escherichia coli K-12. J Bacteriol. 1973 Mar;113(3):1091–1095. doi: 10.1128/jb.113.3.1091-1095.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Sen L. C., Stalker D. M. An Altered aroA Gene Product Confers Resistance to the Herbicide Glyphosate. Science. 1983 Jul 22;221(4608):370–371. doi: 10.1126/science.221.4608.370. [DOI] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemski P., Jr, Stocker B. A. Transduction by bacteriophage P22 in nonsmooth mutants of Salmonella typhimurium. J Bacteriol. 1967 May;93(5):1588–1597. doi: 10.1128/jb.93.5.1588-1597.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiseth S. K., Stocker B. A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981 May 21;291(5812):238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Reichardt K., Botstein D. Inversions and deletions of the Salmonella chromosome generated by the translocatable tetracycline resistance element Tn10. J Mol Biol. 1979 Jan 5;127(1):89–115. doi: 10.1016/0022-2836(79)90461-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan B. D., Buck M., Humphreys J., Griffiths E. Iron-related modification of bacterial transfer RNA. Nucleic Acids Res. 1981 Jun 11;9(11):2629–2640. doi: 10.1093/nar/9.11.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Demerec M., Eisenstark A. Genetic analysis of aromatic mutants of Salmonella typhimurium. Genetics. 1967 Jun;56(2):341–351. doi: 10.1093/genetics/56.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva E. T., Liljeström P., Harayama S. Cosmid cloning and transposon mutagenesis in Salmonella typhimurium using phage lambda vehicles. Mol Gen Genet. 1981;181(2):153–157. doi: 10.1007/BF00268420. [DOI] [PubMed] [Google Scholar]
- Pascal M. C., Casse F., Chippaux M. Localization of pfl gene by transductional study of the gal-aro A segment of the Salmonella typhimurium LT2 chromosome. Mol Gen Genet. 1977 Feb 15;150(3):331–334. doi: 10.1007/BF00268133. [DOI] [PubMed] [Google Scholar]
- Robertsson J. A., Lindberg A. A., Hoiseth S., Stocker B. A. Salmonella typhimurium infection in calves: protection and survival of virulent challenge bacteria after immunization with live or inactivated vaccines. Infect Immun. 1983 Aug;41(2):742–750. doi: 10.1128/iai.41.2.742-750.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. G., Swan J., Kleckner N. Nearly precise excision: a new type of DNA alteration associated with the translocatable element Tn10. Cell. 1979 Apr;16(4):733–738. doi: 10.1016/0092-8674(79)90089-8. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, Edition VI. Microbiol Rev. 1983 Sep;47(3):410–453. doi: 10.1128/mr.47.3.410-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M. B., Roth J. R. Genetic methods for analysis and manipulation of inversion mutations in bacteria. Genetics. 1983 Nov;105(3):517–537. doi: 10.1093/genetics/105.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Dempsey W. B. 3-hydroxypyruvate substitutes for pyridoxine in serC mutants of Escherichia coli K-12. J Bacteriol. 1978 Jun;134(3):944–949. doi: 10.1128/jb.134.3.944-949.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. P., Reina-Guerra M., Hoiseth S. K., Stocker B. A., Habasha F., Johnson E., Merritt F. Aromatic-dependent Salmonella typhimurium as modified live vaccines for calves. Am J Vet Res. 1984 Jan;45(1):59–66. [PubMed] [Google Scholar]
- Stocker B. A., Nurminen M., Mäkelä P. H. Mutants defective in the 33K outer membrane protein of Salmonella typhimurium. J Bacteriol. 1979 Aug;139(2):376–383. doi: 10.1128/jb.139.2.376-383.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


