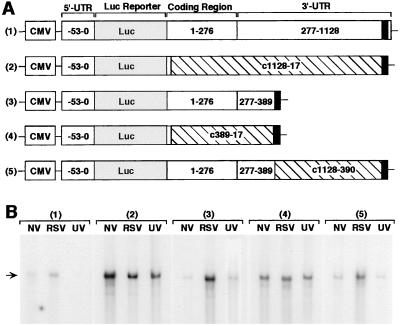
Figure 4.
Basal destabilizing and virus-induced stabilizing activity of RANTES mRNA sequence. (A) The construct designs are depicted for five reporter plasmids: 1) pGL3-CMV-RANTES(-53–1160)-luc, 2) pGL3-CMV-RANTES(-53–10/c1128–17)-luc, 3) pGL3-CMV-RANTES(-53–389)-luc, 4) pGL3-CMV-RANTES(-53–10/c389–17)-luc, and 5) pGL3-CMV-RANTES(-53–389/c1128–390)-luc. Plasmid components include: CMV promoter, P. pyralis luciferase (luc) reporter (309 bp), and RANTES 5′ UTR (nucleotide −53 to transcription start site), coding region (nucleotides 1–276), and 3′ UTR (nucleotides 277-1160) including the polyadenylation signal (nucleotides 1129–1154 represented as darkened box), and reverse-complement of RANTES sequence (denoted by c and crosshatched boxes). (B) hTBE cell monolayers were transfected with one of the six reporter plasmids and a control reporter plasmid pRL-CMV expressing R. reniformis luciferase, and 18–42 h later the transfected cells were treated with no virus (NV), RSV MOI 1.0 (RSV), or UV-inactivated RSV (UV) for an additional 24 h. For each condition, the level of luciferase reporter mRNA was determined by a single-tube RNase protection assay in which total cellular RNA (30 μg/condition) was isolated and hybridized with 32P-labeled RNA luc probe, and protection from RNase degradation was detected by 8% nondenaturing PAGE and autoradiography. Arrow indicates position of protected luciferase mRNA fragment. Transfection efficiency/expression (based on R. reniformis luciferase activity and plasmid copy number) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels (based on protection by an RNA probe for GAPDH) was similar for all treatment conditions (data not shown). Results are representative of five experiments.