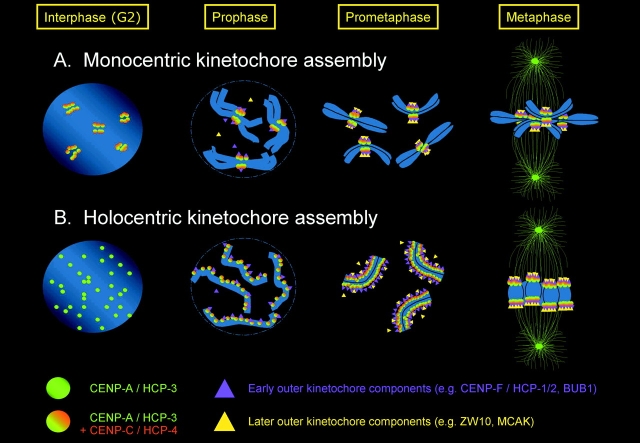
Figure 1.
Assembly of kinetochores on monocentric and holocentric chromosomes. This schematic diagram illustrates the major events of kinetochore assembly, based on information from a body of work. CENP-A/HCP-3 decorates centromeres throughout the cell cycle. In monocentric organisms (A), CENP-A staining appears as distinct foci corresponding to the number of chromosomes. These foci recruit other components, including checkpoint proteins, motors, and structural elements, during prophase to build the mitotic kinetochore. The order of assembly of many components is not yet known. In C. elegans interphase nuclei (B), HCP-3 staining reveals a multitude of tiny foci. However, by the time worm chromosomes can be seen as individual entities at prophase, a single ribbon of semicontiguous HCP-3 staining is evident along one edge of each pair of sisters. HCP-4 (CENP-C) and HCP-1 (CENP-F) appear to be recruited during prophase, as does BUB-1. MCAK loads slightly later than BUB-1. As the chromosomes congress to the metaphase plate, the single ribbon of staining splits into two ribbons that separate to opposing faces of the paired sisters. Between this stage and the alignment of the chromosomes at the metaphase plate, each ribbon of kinetochore proteins condenses dramatically to form a dot ∼0.2–0.5 μm in diameter, similar in size to a mammalian centromere. A notable difference between mono- and holocentric kinetochores is that “splitting” of moncentric kinetochores into two sister foci is already evident by G2, when centromeres appear as “double dots.” In contrast, in C. elegans there is no cytological indication of the duality of the replicated chromosome until late in mitotic prophase. Why this splitting is apparent later in worms is unknown.