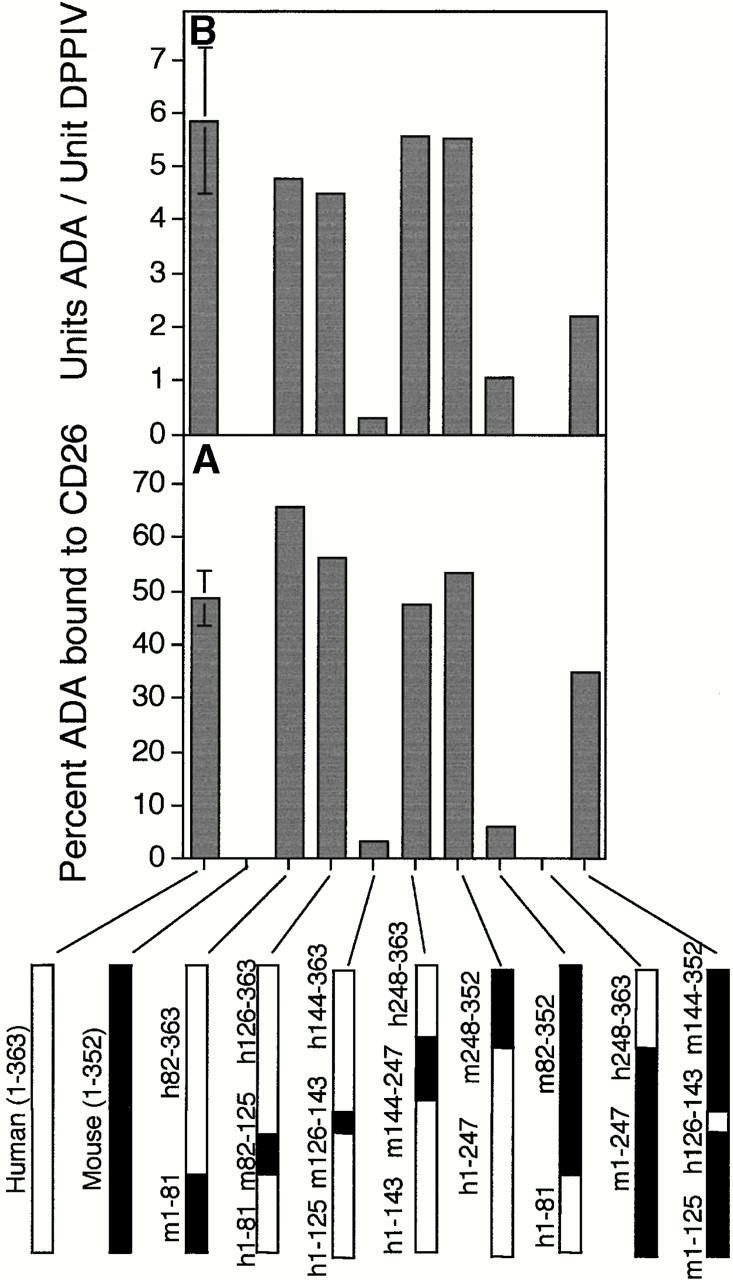
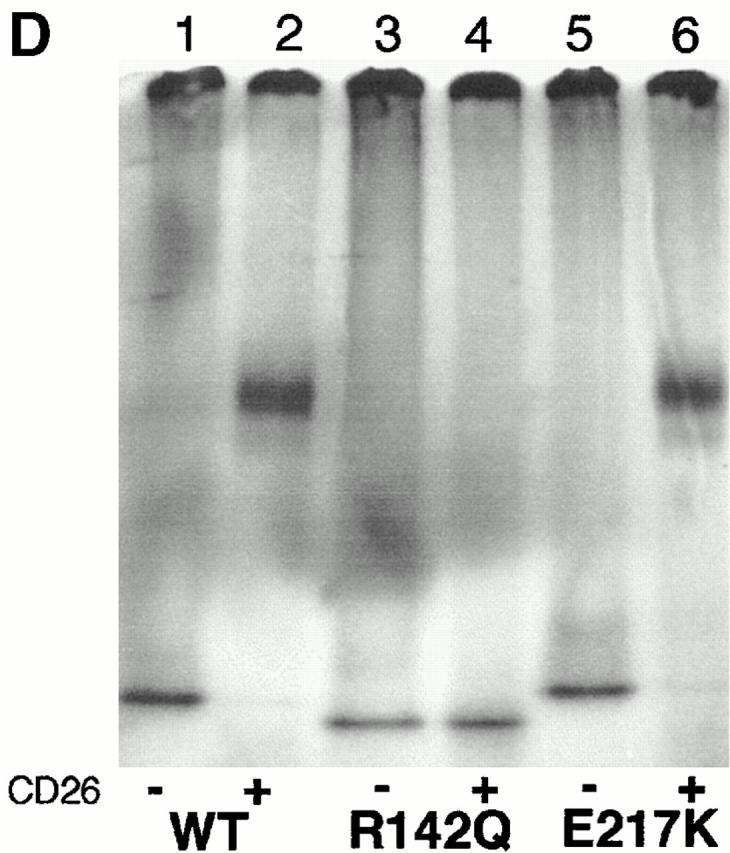
Figure 2.
Binding of rabbit CD26 by human–mouse ADA hybrids and ADA point mutants. (A and B) FPLC assay of human–mouse ADA hybrids. Hybrid structures are shown as bars in which white indicates human (h) and black indicates mouse (m) ADA amino acid residues (numbered beside bars). Gray bars in A and B show the percentage of input ADA activity associated with DPPIV activity (A), or the units of ADA bound per unit of DPPIV (B). Error bars (SD) shown for wild-type human ADA (A–C) and the R142Q mutant (C) are based on five and four standard binding experiments (see Materials and Methods), respectively. (C) FPLC assay of human (Hum) and mouse (Mus) ADA point mutants. Black bars are mutants found in ADA-deficient individuals; gray bars are mutants prepared for this study from human or mouse wild-type cDNA, as indicated. Standard FPLC assay conditions were used for screening the hybrids and point mutants, except for the R149Q, A215T, P126Q mutants from ADA-deficient individuals. In those cases, a lower amount of ADA activity was used, accounting for a greater percentage of input ADA activity becoming associated with CD26/DPPIV than was found with wild-type human ADA. (D) Nondenaturing PAGE assay. Lane 1, wild-type (WT) human ADA; lane 2, wild-type human ADA plus rabbit CD26; lane 3, R142Q human ADA mutant; lane 4, R142Q plus rabbit CD26; lane 5, E217K human ADA mutant; lane 6, E217K plus rabbit CD26.