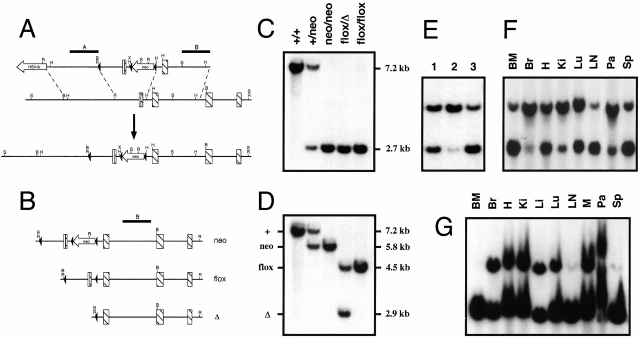
Figure 1.
Targeting strategy and conditional deletion of the vcam-1flox allele. (A) VCAM-1 knock-in mice (bottom locus map) contain Cre recombinase sites of recombination (loxP sites; black arrowheads). The vcam-1 coding sequence exons are depicted as striped bars, with the 5′ untranslated region as a white bar. Probes A and B are shown above the top map line as black bars. Endonuclease sites shown are BamHI (B), EcoRI (R), HindIII (H), SphI (S), and XhoI (X). (B) The vcam-1neo allele (neo) is shown along with two of the possible outcomes of Cre recombinase–mediated deletion, the vcam-1flox allele (flox) and the vcam-1 Δ allele (Δ). (C and D) BamHI Southern blot analysis of tail DNA using probes A and B, respectively. Deletion of the vcam-1neo allele to generate the vcam-1flox allele (flox) and the vcam-1 Δ allele (Δ) in whole mice was achieved by interbreeding with the splicer transgene. (E) Southern blot analysis of mouse tail DNA as in D. A vcam-1flox/Δ mouse (track 1) is shown alongside two TIE2Cre+ mice born of vcam-1flox/flox parents, but with the TIE2Cre+ parent being the father (track 2) or mother (tracks 1 and 3). (F and G) Southern blot analysis as in D of genomic DNA from a vcam-1flox/flox/TIE2Cre+ mouse and a vcam-1flox/Δ/TIE2Cre+ mouse, respectively. Tissues analyzed were: BM; Br, brain (cortex); H, heart; Ki, kidney; Li, liver; Lu, lung; LNs; M, muscle (thigh); Pa, pancreas; and Sp, spleen.