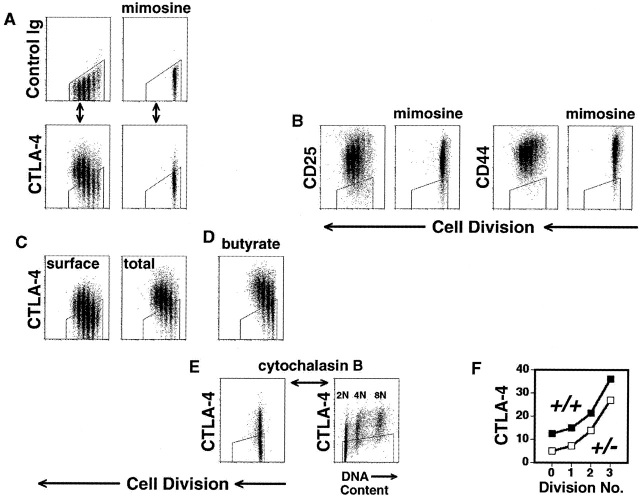
Figure 4.
Cell cycle–coupled induction of CTLA-4. (A–E) C57BL/6, CD8-depleted, CFSE-labeled splenocytes were stimulated with anti-CD3 (2.0 μg/ml) for 3 d. All flow cytometric plots depict only live-gated, CD4+ events. Polygonal gates are drawn around the upper limit of background staining, as illustrated in part A. (A) After culture in the absence (left) or presence (right) of mimosine (300 μM), cells were fixed, permeabilized, and stained (see Materials and Methods) with anti-CD4 mAb and either fluorochrome-conjugated hamster control mAb (top row) or hamster anti–mouse CTLA-4 mAb (lower row), before analysis of cell division (x-axis) versus control staining (top) or total cellular CTLA-4 expression (bottom) (y-axis). (B) Cells were washed and stained (without prior fixation or permeabilization) with either anti-CD25, anti-CD44, or control mAb, before analysis of cell division (x-axis) versus surface expression (y-axis) of CD25 (left panels) and CD44 (right panels). (C) Cells were washed, fixed and either stained directly (“surface,” left panel) or permeabilized before staining (“total,” right panel) as above. (D) In the same experiment as C, a group of cells was stimulated in the presence of sodium butyrate (600 μM), to inhibit histone deacetylases, and analyzed for total cellular CTLA-4 expression (y-axis). (E) Cytochalasin B (3.5 μg/ml) was used to prevent cytokinesis in stimulated cells (left panel) before analysis of both DNA content (x-axis, right panel) and CTLA-4 expression (y-axis). (F) Splenocytes from 4 week-old BALB/c CTLA-4+/+ (filled symbols) and CTLA-4+/− (open symbols) littermate mice were stimulated and analyzed as in part A. Geometric mean fluorescence intensity of CTLA-4 expression (y-axis) for each cell generation (x-axis) is displayed. All experiments were performed at least twice.