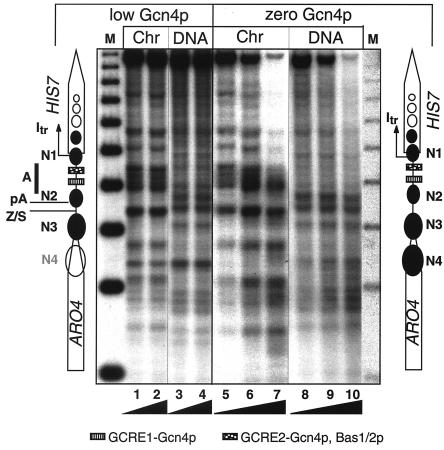
FIG. 2.
Chromatin structure at the ARO4-HIS7 intergenic region. Mnase protection experiments are shown with chromatin from strains FY1353 (wild-type GCN4) or RH1381 (gcn4Δ). Cells were cultivated in the absence of exogenous adenine. Control Mnase digests of naked DNA are shown in lanes 3, 4, 8, 9, and 10. Lanes M on the far left and right show a 256-bp DNA-ladder (42). Black ramps at the bottom indicate increasing amounts of Mnase and exactly represent 20 and 40 U for lanes 1 and 2; 20, 40, and 60 U for lanes 5, 6, and 7; 4 and 8 U for lanes 3 and 4; and 4, 8, and 12 U for lanes 8, 9, and 10, respectively. The diagrams on the left and right show the location of the open reading frames of ARO4 and HIS7. Protected regions are shown as black ovals N1-N3 (here referred to as positioned nucleosomes). The binding sites for Gcn4p and Bas1p/Bas2p are indicated at the bottom. The site of HIS7 transcriptional initiation is indicated by Itr. pA, poly(A) addition site of the ARO4 mRNA; Z/S, Zaret/Sherman element required for efficient ARO4 mRNA 3′-end processing.