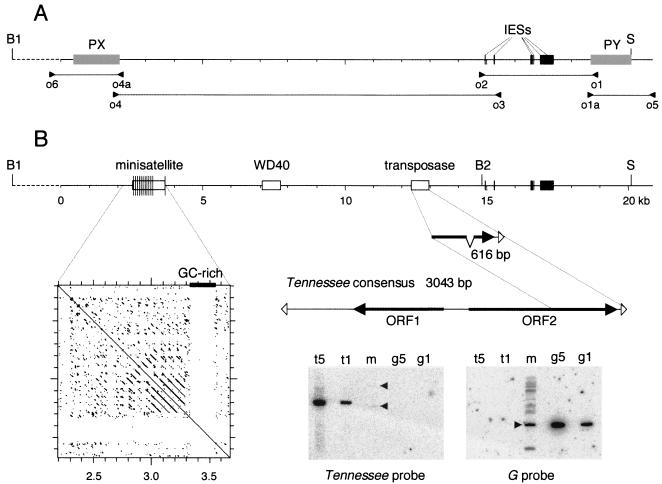
FIG. 3.
Germ line sequence of the chromosome fragmentation region. (A) PCR amplification of micronuclear DNA. The positions of primers are indicated by arrowheads. Primers o1 and o1a and primers o4 and o4a are located within the PY and PX sequences, repectively (gray boxes). Micronuclear specificity was achieved by using DNA from ΔPY clones for the o1-o2 and o1a-o5 PCRs and DNA from ΔPX clones for the o3-o4 and o4a-o6 PCRs. In addition, primer o3 was derived from one of the six IESs, 26 to 468 bp long (black boxes), that were identified in the o1-o2 micronuclear product. (B) Features of the germ line sequence. The 20,816-bp sequenced portion (solid line) begins 1.7 kb after BamHI site B1 and ends 0.8 kb after SacI site S. White boxes indicate the positions of the minisatellite and GC-rich sequence, WD40-related sequence, and truncated transposon. Thin vertical lines in the minisatellite represent individual repeats. The dot plot shows a self-comparison matrix of the minisatellite and GC-rich sequence (DNA Strider stringency 15; window 23). The 616-bp sequence of the truncated Tennessee copy is compared with the 3,043-bp consensus sequence derived from the alignment of eight different copies. ORFs are indicated by filled arrows, and terminal 31-bp IRs are indicated by open arrowheads. The Southern blot at the bottom was hybridized successively with a 677-bp probe from the right end of the Tennessee transposon (left panel) and, as a control, with a 1.6-kb EcoRI fragment from the G surface antigen gene (right panel). Lanes t5 and t1 contain 150 and 30 pg of a 3-kb complete Tennessee copy, respectively; these amounts correspond to five copies and one copy per haploid genome in 1 μg of the ∼100-Mb macronuclear genome. Lanes m contain 1 μg of EcoRI-digested DNA from the reference clone. Only two faint bands were revealed by the Tennessee probe in lane m (arrowheads), indicating that two copies of the transposon are partially maintained in the macronucleus, at much less than one copy per haploid genome. Lanes g5 and g1 contain 80 and 16 pg of the 1.6-kb EcoRI fragment from the G surface antigen gene, respectively; these amounts correspond to five copies and one copy per haploid genome. The 1.6-kb fragment revealed by the G surface antigen gene probe in lane m (arrowhead) indicates that the G surface antigen gene is present at one copy per haploid genome in macronuclear DNA; weaker bands arise from paralogous genes that cross-hybridize with the probe under the low-stringency conditions used.