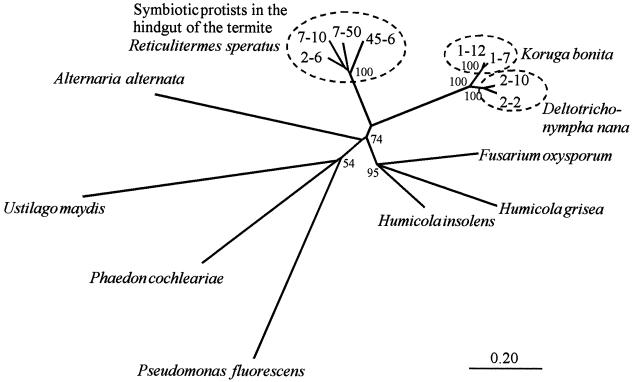
FIG. 7.
Unrooted phylogenetic tree of GHF45 members based on the amino acid sequence alignment of the catalytic domains (shown in Fig. 6) without putative signal sequences. All gaps in the alignment were omitted, leaving 207 amino acid positions for phylogenetic analysis. Bootstrap values (100 replicates) are indicated at their respective branching points. Bar, 0.20 amino acid substitution.