Abstract
With the goal of the genetic characterization of the d-xylose pathway in Hypocrea jecorina (anamorph: Trichoderma reesei), we cloned the xdh1 gene, encoding NAD-xylitol dehydrogenase, which catalyzes the second step of fungal d-xylose catabolism. This gene encodes a 363-amino-acid protein which has a mass of 38 kDa, belongs to the zinc-containing alcohol dehydrogenase family, exhibits high sequence identity to the published sequences of xylitol dehydrogenases from yeast origins, but contains a second, additional binding site for Zn2+. The enzyme catalyzed the NAD-dependent oxidation of xylitol and d-sorbitol and the NADH-dependent reduction of d-xylulose and d-fructose. No activity was observed with NADP, l-arabinose, or l-arabinitol. A single 1.4-kb transcript was formed during growth on xylan, d-xylose, l-arabinose, l-arabinitol and, at a lower abundance, xylitol, d-galactose, galactitol, and lactose but not on d-glucose and glycerol. xdh1 deletion mutants exhibited 50% reduced growth rates on d-xylose, whereas growth rates on xylitol remained unaltered. These mutants contained 30% of the xylitol dehydrogenase activity of the parent strain, indicating the presence of a second xylitol dehydrogenase. This activity was shown to be due to lad1-encoded l-arabinitol-4-dehydrogenase, because H. jecorina xdh1 lad1 double-deletion strains failed to grow on d-xylose or xylitol. In contrast, lad1 deletion strains of H. jecorina grew normally on these carbon sources. These results show that H. jecorina contains a single xylitol dehydrogenase which is encoded by xdh1 and is involved in the metabolism of d-xylose and that lad1-encoded l-arabinitol-4-dehydrogenase can compensate for it partially in mutants with a loss of xdh1 function.
d-Xylose is a major constituent of plant hemicelluloses, where it forms the β-1,4-xylan backbone of hardwood. β-1,4-Xylans are heteropolysaccharides that have a backbone of β-1,4-linked xylopyranosyl residues and contain side groups such as d-glucuronic acid, l-arabinose, p-coumaric acid, and ferulic acid. They constitute 20 to 35% of the approximately 830 gigatons of annually formed renewable plant biomass (23).
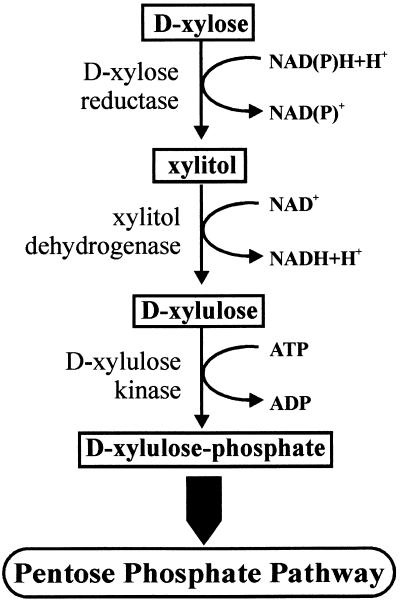
Both prokaryotic and eukaryotic microorganisms can use xylan as a carbon source for growth. The bacterial pathway for d-xylose catabolism is well established. It consists of an isomerase and a kinase that sequentially convert d-xylose to d-xylulose and d-xylulose-5-phosphate, which is an intermediate of the pentose phosphate pathway. This pathway is absent from fungi, where d-xylose instead is subjected to NADPH-linked reduction and NAD-linked oxidation reactions before phosphorylation of d-xylulose occurs (Fig. 1). The enzymes catalyzing the first two steps (aldose reductase [EC 1.1.1.21] and xylitol dehydrogenase [EC 1.1.1.9]) have been characterized mainly from yeasts. They are nonspecific and can use other sugars in addition to d-xylose and xylitol, respectively, at approximately the same rates. The genes encoding the enzymes involved in fungal d-xylose catabolism have also been cloned from different yeasts (for a review, see reference 10) and, in part (i.e., aldose reductase and xylulose-5-phosphate kinase), from Aspergillus niger (9, 25). Further, cloning of a gene encoding a xylitol dehydrogenase from Hypocrea jecorina has been reported, but neither its nucleotide sequence nor its amino acid sequence has been made available (27).
FIG. 1.
Fungal d-xylose pathway.
A genetic analysis of the d-xylose-metabolizing pathway in yeasts showed that aldose reductase and xylitol dehydrogenase are indeed essential for d-xylose degradation. In contrast, an aldose reductase knockout mutant of A. niger was still able to grow on d-xylose, although at a reduced rate (9), suggesting that multiple enzymes are involved in the first step of the d-xylose catabolic pathway in filamentous fungi.
Xylan breakdown by the ascomycete H. jecorina (anamorph: Trichoderma reesei) has received the strongest interest because of the application of the corresponding xylanases in the pulp and paper industry and the food industry. The extracellular addition of intermediates of the d-xylose metabolic pathway leads to differential expression of the two major xylanase genes (14), stressing the need to understand this pathway in more detail for the metabolic engineering of xylanase formation in this fungus. As a first step toward the genetic engineering of the d-xylose catabolic pathway in H. jecorina, we attempted to identify the genes and proteins involved.
Here we report the cloning of the xdh1 gene, encoding NAD-xylitol dehydrogenase of H. jecorina. We show that deletion of the gene partially affects the growth of H. jecorina on d-xylose and that l-arabinitol-4-dehydrogenase (encoded by lad1) partially compensates for the loss of xdh1 function under these conditions. However, we also show that lad1 is not involved in d-xylose metabolism in a background in which xdh1 is functional; this result identifies the lad1 bypass as a rescue mechanism only.
MATERIALS AND METHODS
Strains and culture conditions.
The H. jecorina parent strains used in this study were QM9414 (ATCC 26921) and the pyr4-negative mutant TU-6 (ATCC MYA-256) (7). All strains were maintained on malt extract agar, and auxotrophic strains were supplemented with uridine (10 mM). Strains were grown in 1-liter Erlenmeyer flasks on a rotary shaker (250 rpm) at 30°C with 250 ml of the medium described by Mandels and Andreotti (16) and containing various carbon sources at final concentrations of 10 g/liter.
For analysis of the effects of different carbon sources on gene expression, the different H. jecorina strains were pregrown on 1% (wt/vol) glycerol (20 h), the mycelia were harvested by filtration and washed with tap water, equal amounts of mycelia (1 ± 0.2 g [wet weight]/liter [mean and standard deviation]) were transferred to flasks containing the different carbon sources (1% [wt/vol]), and cultivation was continued.
Escherichia coli strains ER1647 and BM25.8 (Novagen, Madison, Wis.) were used for genomic library screening, strains XL-1 Blue and XLOLR (Stratagene, La Jolla, Calif.) were used for cDNA library screening, and strain JM109 (Promega, Madison, Wis.) was used for plasmid propagation.
Determination of fungal growth.
To determine hyphal growth on agar plates, a small piece of agar was placed in the center of an 11-cm plate, and the increase in colony diameter was measured twice daily. To measure growth in submerged cultures, the increase in biomass dry weight was recorded. To this end, mycelia were harvested after appropriate times, washed extensively with tap water and then distilled water, and dried to a constant weight.
Preparation of a cell extract.
To prepare a cell extract for enzyme activity assays, H. jecorina strains QM9414 and ΔXDH1 were grown for 24 h on the medium described by Mandels and Andreotti (16). The mycelia were then harvested by filtration through a precooled linen cloth, washed with cold tap water, ground to a fine powder under liquid nitrogen, and homogenized by sonication of a concentrated mycelial suspension (1 g [wet weight] per 2.5 ml of buffer A [0.1 M Tris-HCl {pH 7.5}, 1 mM EDTA, 5 mM β-mercaptoethanol]) 10 times for 30 s each time at 2°C. The resulting homogenate was centrifuged at 10,000 × g for 20 min at 4°C. The supernatant, which had a protein content of between 8 and 15 mg/ml, was used as a cell extract.
Enzyme assay.
Xylitol dehydrogenase activity was assayed by the procedure of Chakravorty and Horecker (3) with 0.1 M glycine buffer (pH 8.6) and 0.1 M glycylglycine (pH 7.0) for the forward and reverse reactions, respectively. Enzyme (cell extract or purified glutathione S-transferase [GST]-Xdh1 fusion) was added in amounts sufficient to produce a change in the A340 of between 0.02 and 0.1/min. The reaction was started by the addition of substrate. Activity was expressed as units, 1 U corresponding to the conversion of 1 μmol of substrate per min, and reported as specific activity (units per milligram of protein). The protein concentration was determined by using a protein assay from Bio-Rad Laboratories, Munich, Germany.
SDS-PAGE.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed with 10% polyacrylamide gels as described by Ausubel et al. (2). Gels were stained with Coomassie blue.
Extraction and quantification of mycelial xylitol.
To measure the intracellular xylitol concentration, mycelia of H. jecorina were harvested, washed with cold tap water, and resuspended in 1 ml of distilled water. The suspension was then snap-frozen at −75°C for 1 h, thawed, boiled for 10 s, and finally homogenized in a precooled Potter-Elvehjem glass homogenizer. The homogenate was centrifuged (10,000 × g, 4°C, 10 min), and the xylitol concentration in the supernatant was quantified by high-pressure liquid chromatography with an H+ exchange column (Bio-Rad Aminex HPX-H+), with 10 mM H2SO4 at 55°C as the mobile phase, and with isocratic elution. Compounds were detected by a refractive index detector. The data were averaged and deviated by not more than 3%. For calculation of the intracellular xylitol concentration, 1 g of mycelial dry weight was assumed to be equivalent to a 2.4-ml intracellular volume (22) without consideration of intracellular compartmentalization.
Cloning of the H. jecorina xdh1 gene.
An alignment of different xylitol dehydrogenases from the National Center for Biotechnology Information data bank revealed the conserved amino acid sequences TGICGSDVH and GHYVQGGM to be potentially suitable for amplifying a corresponding fragment of H. jecorina. Consequently, the primers xdhfor1 (5′-ACCGGCATCTGCGGCTCCGATGTCC-3′) and xdhrev2 (5′-CCGGTGATGCAGGTCCCGCCATAC-3′) were deduced directly from the respective nucleotide sequence of a hypothetical protein (NCU00891.1) in the Neurospora crassa genome database (http://www.genome.wi.mit.edu/annotation/fungi/neurospora) which shows a high level of similarity to xylitol dehydrogenases. The conditions used for amplification with these primers were as follows: 100 ng of H. jecorina QM9414 genomic DNA as a template in a total volume of 50 μl in an automated temperature cycling device (Biotron; Biometra, Göttingen, Germany); a reaction mixture containing 2.5 mM MgCl2, 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 0.1% (vol/vol) Triton X-100, 0.4 μM each primer, 0.2 mM each deoxynucleoside triphosphate, and 0.5 U of Taq polymerase (Promega); and an amplification program consisting of 1 min of initial denaturation (94°C), 30 cycles of amplification (1 min at 94°C, 1 min at 54°C, and 1 min at 74°C), and 7 min of final extension (74°C). These primers allowed the amplification of a 650-bp fragment, which was isolated and used to screen a genomic λ BlueSTAR library (Novagen) of H. jecorina QM9414. The corresponding xdh1 gene was located on a 2.5-kb SacII fragment, ligated to pBluescript SK(+), resulting in pXDH, and sequenced by means of a LI-cor 4200 automatic sequencer (LI-cor Inc., Lincoln, Nebr.). To confirm the positions of putative introns, a complete xdh1 cDNA fragment, isolated from a λ HybriZAP cDNA library (Stratagene) of H. jecorina QM9414 grown on d-xylose, was sequenced.
To amplify a 1.2-kb fragment of H. jecorina lad1 by PCR, primers lad1fw (5′-ACAGCTCGCCATGTCGCCTTC-3′) and lad1rv (5′-GCACCTCACTCTCAATCCAGGC-3′) were derived from the published H. jecorina lad1 sequence (18). Standard conditions were used for amplification. The corresponding lad1 gene was located on ∼11-kb NotI λ genomic clone H1 and partially sequenced.
Sequence analysis.
The 2.5-kb fragment was analyzed by using BLAST programs (1), and a multiple sequence alignment was done by using MultiAlin (4). Consensus binding sequences in the xdh1 and lad1 5′ regions were identified manually.
Nucleic acid isolation and hybridization.
Fungal mycelia were harvested by filtration, washed with tap water, frozen, and ground in liquid nitrogen. Nucleic acids (DNA and total RNA) were extracted as described previously (21). Standard methods (20) were used for electrophoresis, blotting, and hybridization of nucleic acids.
Probes used for Northern hybridization were a 1.9-kb Acc65I act1 fragment (actin encoding), the 1.2-kb lad1 fragment (see above), and a 1.4-kb xdh1 cDNA fragment. The relative abundances of transcripts were determined by densitometric measurements of autoradiographs derived from different exposure times (only values with a linear correlation [r > 0.9] were used).
Overexpression of xdh1 in E. coli.
To obtain purified H. jecorina xylitol dehydrogenase, the full-length xdh1 cDNA was overexpressed as a GST fusion in E. coli. To this end, the xdh1 coding region was PCR amplified from the cDNA clone with primers GEX-XDHfwd (5′-CTGCTGGATCCATGGCGACTCAAACGATC-3′) and GEX-XDHrev (5′-AGGGCGGCGGCCGCTTACACCTTCTCGTTG-3′). PCR amplification was performed with Pfu polymerase (Promega) by using an initial denaturation cycle of 45 s at 94°C, 28 cycles of amplification (45 s at 94°C, 45 s at 50°C, and 3 min at 72°C), and a final extension step of 10 min at 72°C. The amplicon was cut with BamHI and NotI and cloned into pGEX4-2T (Amersham Biosciences, Vienna, Austria). After verification by sequencing, the GST-Xdh1 fusion protein was overexpressed in E. coli BL21 (Stratagene) and purified by using glutathione-Sepharose 4B (Amersham Biosciences) according to the manufacturer's protocol. For storage at −80 or −20°C, 20% (vol/vol) sterile glycerol was added.
Construction of H. jecorina xdh1, lad1, and xdh1 lad1 knockout mutants.
To construct an xdh1 knockout vector, a genomic fragment containing 3 kb upstream and 4 kb downstream of the xdh1 coding region was isolated from the λ phage clone described above, and the xdh1 coding region was replaced by the hygromycin resistance-conferring expression cassette from pRLMex30 (15). To this end, a 3-kb ApaI-ClaI fragment of the xdh1 upstream region was cloned into pBluescript SK(+), the 2.8-kb XhoI-HindIII hygromycin resistance cassette was cloned into the resulting vector, and finally a 2.8-kb ApaI-SalI fragment of the xdh1 downstream region was added to this vector to result in the final deletion vector pΔXDH. Transformation of H. jecorina QM9414 was done by the protocol of Gruber et al. (7) with an 8.0-kb Acc65I-BstXI fragment of pΔXDH.
To construct a lad1 knockout strain, the lad1 coding region was replaced by the H. jecorina pyr4 marker (6). To this end, the 2.7-kb SalI pyr4 fragment was cloned between a 1-kb BamHI-MluI upstream fragment and a 2-kb EcoRV-NcoI downstream fragment of lad1 in pBluescript SK(+), resulting in the final deletion vector pΔLAD1. For transformation of H. jecorina TU-6, a 5.7-kb ApaI-XbaI fragment of pΔLAD1 was used.
To obtain strains with deletions of both dehydrogenases, H. jecorina Δlad1 was transformed with the 8.0-kb Acc65I-BstXI fragment of pΔXDH.
Nucleotide sequence accession numbers.
The DNA sequences assembled here were deposited in GenBank under the following accession numbers: xdh1, AF428150; and lad1, AY225444.
RESULTS
Cloning and characterization of H. jecorina xdh1 and its encoded protein.
Based on the PCR approach outlined above, a 650-bp H. jecorina xylitol dehydrogenase fragment was amplified and used to isolate a 2.5-kb genomic subclone which included the complete structural xdh1 gene. The results of Southern analysis of chromosomal DNA digested with different restriction nucleases are consistent with the occurrence of a single xdh1 gene in the H. jecorina genome (data not shown). Nucleotide sequence analysis revealed an open reading frame (ORF) of 1,210 bp, interrupted by a single intron of 118 bp, encoding a 363-amino-acid protein with a calculated mass of 38 kDa. An analysis of the amino acid sequence with PROSITE (http://www.expasy.ch/prosite) identified the protein as a member of the zinc-containing alcohol dehydrogenase family, showing highest sequence identity to the xylitol dehydrogenase of Candida sp. strain HA167 (former Galactocandida mastodermitis). When the Xdh1 protein sequence was compared with the Neurospora crassa database, four ORFs showed significant degrees of similarity (E values of 1e−160, 1e−60, 1e−48, and 1e−44). The highest similarity was found with hypothetical protein NCU00891.1, from which the primers were developed. The second highest similarity was obtained with NCU00643.1. However, submitting NCU00643.1 to a BLASTP search revealed that it is highly similar to l-arabinitol-2-dehydrogenase of H. jecorina (18). The two even less similar proteins (NCU07022.1 and NCU01905.1) could not be identified with reasonable certainty.
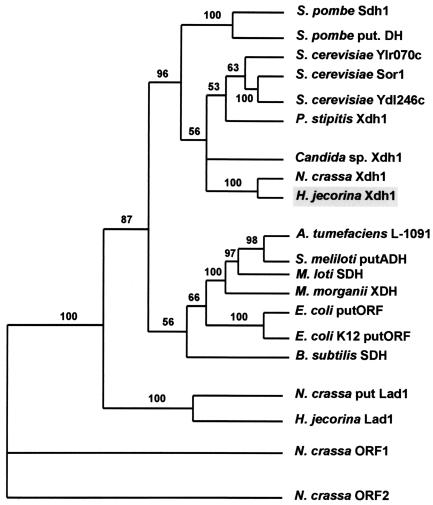
A parsimony analysis of the amino acid sequence of H. jecorina Xdh1 and those of several other prokaryotic and eukaryotic proteins to which it showed the highest similarity in a BLAST search revealed that Xdh1 and hypothetical protein NCU00891.1 of N. crassa formed a strongly supported terminal branch within a dichotomous cluster containing all other yeast xylitol and d-sorbitol dehydrogenases and for which two d-sorbitol dehydrogenases of Schizosaccharomyces pombe represented a basal ancestor (Fig. 2). Seven bacterial alcohol and d-sorbitol dehydrogenases formed a sister clade to this cluster. The H. jecorina and N. crassa l-arabinitol dehydrogenases formed a clearly different cluster. These data provide evidence that the protein encoded by H. jecorina xdh1 clearly is a member of the fungal xylitol and d-sorbitol dehydrogenase family and, in addition, that only one such member is present in the Neurospora genome sequence database.
FIG. 2.
Phylogenetic tree inferred by parsimony analysis of the amino acid sequences of xylitol dehydrogenases and d-sorbitol dehydrogenases. The tree shown is the single most parsimonious tree. The numbers given over selected branches indicate the percentages of 500 bootstrap-resampled data sets supporting the clade to the right of the branch and are given only for values exceeding 50. Proteins used in the tree were as follows: S. pombe Sdh1 (P36624), S. pombe putative dehydrogenase (put. DH) (S38345), Saccharomyces cerevisiae Ylr070c (NP_013171.1), S. cerevisiae Sor1 (NP_012693.1), S. cerevisiae Ydl246c (NP_010035.1), Pichia stipitis Xdh1 (P22144), Candida sp. Xdh1 (AAC24597.1), N. crassa Xdh1 (NCU00891.1), H. jecorina Xdh1 (AF428150), Agrobacterium tumefaciens L-1091 (NP_356336.1), Sinorhizobium meliloti putative alcohol dehydrogenase (putADH) (NP_386632.1), Mesorhizobium loti d-sorbitol dehydrogenase (SDH) (NP_105675.1), Morganella morganii xylitol dehydrogenase (XDH) (AAA25324.1), E. coli putative ORF (putORF) product (NP_288210.1), E. coli K-12 putative ORF product (NP_416288.1), Bacillus subtilis SDH (NP_388496.1), N. crassa putative l-arabinitol-2-dehydrogenase (put Lad1) (NCU00643.1), H. jecorina l-arabinitol-2-dehydrogenase (Lad1) (AAL08944.1), N. crassa ORF1 product (NCU07022.1), and N. crassa ORF2 product (NCU01905.1).
Figure 3 shows an alignment of the amino acid sequence of H. jecorina xylitol dehydrogenase with those of its closest yeast neighbors: the polyol and coenzyme binding sites as well as the zinc binding sites are well conserved, but there is generally only poor conservation outside these areas. It is interesting that H. jecorina Xdh1 and N. crassa Xdh1 (NCU00891.1)—in contrast to the yeast xylitol dehydrogenases—have two predicted binding sites for Zn2+ instead of one: one site, consisting of C50, H75, and E161, which is typical for d-sorbitol and xylitol dehydrogenases, and a second site, comprising C105, C108, C111, and C119, which is typical for alcohol dehydrogenase and which is not found in the other xylitol dehydrogenases described so far from yeast origins.
FIG. 3.
Alignment of H. jecorina xylitol dehydrogenase with other xylitol and d-sorbitol dehydrogenases. Dehydogenases were as follows: NcXdh1, N. crassa Xdh1 (NCU00891.1); HjXdh1, H. jecorina Xdh1; CaXdh1, Candida sp. strain HA167 Xdh1 (AAC24597.1); ScSor1, S. cerevisiae Sor1 (NP_012693.1); ScXdh1, S. cerevisiae Xhd1 (NP_013171.1); PsXdh1, P. stipitis Xdh1 (P22144); and SpSdh1, S. pombe Sdh1 (P36624). Diamonds indicate the first zinc binding site (C50, H75, and E161), and asterisks indicate the second zinc binding site (C105, C108, C111, and C119). The dashes over the sequence indicate the zinc-containing alcohol dehydrogenase signature, and the plus signs over the sequence indicate an NADbinding site. Residues in white on a black background are conserved in at least 90% of the proteins; residues in white on a grey background are conserved in 40%.
Regulation of xdh1 gene expression.
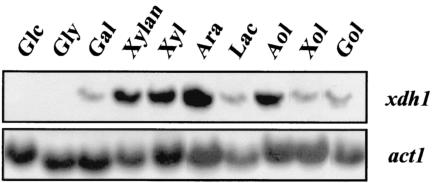
The transcriptional regulation of xdh1 was studied by Northern analysis: during growth on d-xylose or xylan, H. jecorina accumulated a single, 1.4-kb xdh1 transcript (Fig. 4). An xdh1 transcript accumulated on l-arabinitol to an abundance similar to that on d-xylose or xylan, whereas its abundance was found to be higher on l-arabinose but significantly lower on d-galactose, galactitol, lactose, or xylitol. Generally, the xdh1 transcript was most abundant in young cultures, and its abundance was sharply decreased during further cultivation (data not shown). No xdh1 transcript could be detected during growth on d-glucose or glycerol.
FIG. 4.
Induction of xdh1 transcription in H. jecorina by different carbon sources. The results of a Northern analysis of xdh1 transcript levels during growth on various carbon sources are shown. Samples were obtained 6 h after transfer from a glycerol culture to cultures with the carbon sources shown. For the carbon sources lactose and xylan, samples were obtained after 24 h of batch growth (conidial inoculum). Abbreviations: Glc, d-glucose; Gly, glycerol; Gal, d-galactose; Xyl, d-xylose; Ara, l-arabinose; Lac, lactose; Aol, l-arabinitol; Xol, xylitol; and Gol, galactitol.
Substrate specificity of Xdh1.
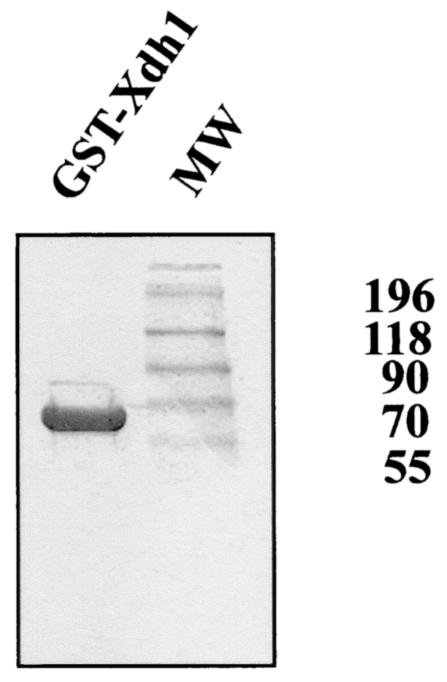
H. jecorina Xdh1 was overexpressed in E. coli as a fusion to GST, purified to physical homogeneity (Fig. 5), and used to investigate the substrate specificity of the enzyme. The enzyme was active with xylitol and d-sorbitol in the forward reaction and with d-xylulose and d-fructose in the reverse reaction. No activity was observed with l-arabinose or l-arabinitol as a substrate. NAD or NADH was exclusively required as a coenzyme; NADP or NADPH yielded less then 5% activity (Table 1). We also tested whether Mg2+ was necessary and found full activity in the absence of Mg2+. These data are largely consistent with those reported for xylitol dehydrogenase from the yeast Candida (13) and suggest that the two enzymes—despite having different numbers of predicted zinc binding sites—have similar substrate specificities.
FIG. 5.
SDS-PAGE of purified GST-xylitol dehydrogenase fusion protein. Ten micrograms of protein was loaded on the gel, which was stained with Coomassie blue. MW, molecular weight markers (in thousands).
TABLE 1.
Substrate specificity of recombinant Xdh1 of H. jecorinaa
| Substrate | Cosubstrate (mM) | Mean ± SDa
|
|
|---|---|---|---|
| Km (mM) | Vmax (U/mg of protein) | ||
| Xylitol | NAD (0.3) | 25 ± 7 | 0.06 ± 0.02 |
| d-Sorbitol | NAD (0.3) | 24 ± 7 | 0.04 ± 0.01 |
| Galactitol | NAD (0.3) | ND | <0.002 ± 0.001 |
| l-Arabinitol | NAD (0.3) | ND | <0.002 ± 0.001 |
| d-Xylulose | NADH (0.15) | 4.5 ± 0.8 | 0.10 ± 0.03 |
| d-Fructose | NADH (0.15) | 400 ± 120 | 0.15 ± 0.04 |
| d-Galactose | NADH (0.15) | ND | <0.005 ± 0.002 |
| NAD+ | Xylitol (50) | 0.025 ± 0.010 | 0.06 ± 0.002 |
| NADH+ | d-Xylulose (5) | 0.35 ± 0.014 | 0.10 ± 0.03 |
| NADP+ | Xylitol (50) | ND | <0.005 ± 0.001 |
ND, not determined.
xdh1 is involved in but is not essential for growth on d-xylose.
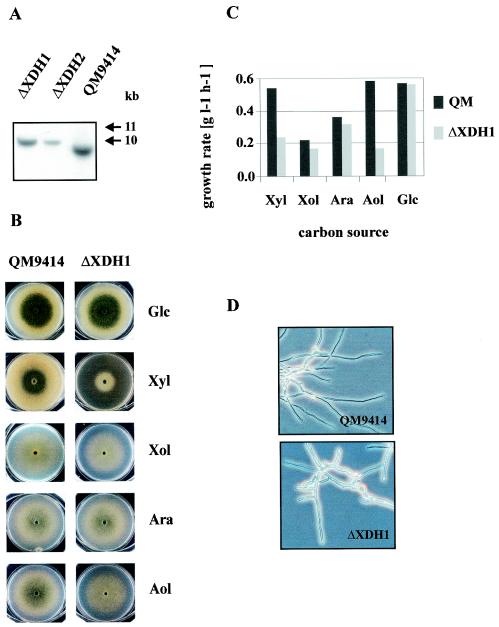
To study whether xylitol dehydrogenase is essential for the metabolism of d-xylose in H. jecorina, a knockout mutant in which the xdh1 coding region was replaced by the E. coli hph gene (encoding hygromycin B phosphotransferase) under the control of H. jecorina expression signals was constructed (15). Several xdh1 disruptants, verified by Southern analysis (Fig. 6A), were obtained. All of them exhibited considerably slower growth on d-xylose but clearly were still able to grow on this carbon source. Interestingly, growth on xylitol—on which the parent strain already grew rather slowly—was unaffected in the deletion mutant (Fig. 6B and C). Consistent with this finding, cell extracts from the deletion mutant still contained xylitol dehydrogenase activity, albeit at a significantly reduced level (Table 2), thus indicating that the xdh1 gene product does not account for all of the xylitol dehydrogenase activity of H. jecorina ΔXDH1. A microscopic examination of the mutant further showed that during growth on d-xylose, the hyphae of the mutant appeared swollen and contained thicker cell walls (Fig. 6D). This morphology correlates with the accumulation of much higher concentrations of xylitol in the hyphae of strain ΔXDH1 than in those of the parent strain (112 versus 55 mM, respectively). During growth on xylitol, on the other hand, the hyphae of strain ΔXDH1 showed no changes in morphology compared with the morphology of the parent strain, consistent with the similar rates of growth of both strains on xylitol as a carbon source.
FIG. 6.
Genetic and physiological characterization of an H. jecorina xdh1 deletion mutant. (A) Southern analysis of xdh1 deletion strains. Genomic DNA was digested with EcoRV and probed with a 2.5-kb SacII fragment of xdh1. Replacement of the xdh1 coding region by the hygromycin resistance-conferring expression cassette in the xdh1 deletion strain leads to an ∼900-bp increase in the size of the 10-kb hybridizing fragment in strain QM9414 to ∼11 kb in the xdh1 deletion strain. (B) Comparison of the growth behaviors of parent strain QM9414 and the xdh1 deletion strain on plates containing d-glucose (Glc), d-xylose (Xyl), xylitol (Xol), l-arabinose (Ara), and l-arabinitol (Aol) as carbon sources. (C) Effect of the xdh1 deletion on biomass production during growth on various carbon sources in shake flasks. QM, QM9414. (D) Microscopic views of strains QM9414 and ΔXDH1 grown on d-xylose.
TABLE 2.
Effect of xdh1 gene deletion in H. jecorina on total polyol dehydrogenase activitiesa
| Substrate (mM) | Mean ± SD U/mg of protein in:
|
|
|---|---|---|
| QM9414 | ΔXDH1 | |
| Xylitol (150) | 0.33 ± 0.05 | 0.11 ± 0.03 |
| d-Sorbitol (150) | 0.27 ± 0.04 | 0.08 ± 0.02 |
| l-Arabinitol (150) | 0.08 ± 0.02 | 0.06 ± 0.02 |
| Galactitol (150) | <0.01 ± 0.005 | <0.01 ± 0.005 |
| d-Fructose (200) | 0.02 ± 0.01 | <0.01 ± 0.005 |
| d-Xylulose (10) | 0.08 ± 0.02 | 0.01 ± 0.005 |
Strains were grown for 24 h on d-xylose as a carbon source, and activities were determined as described in Materials and Methods.
Interestingly, when the xdh1 deletion mutant was tested for growth on other carbon sources, growth on l-arabinitol but not on l-arabinose also was significantly affected: in submerged cultivation, the increase in biomass density was reduced by more than 50%, which is in the same range as the reduction observed with d-xylose (Fig. 6B and C), but no apparent morphology changes were observed.
l-Arabinitol-4-dehydrogenase is responsible for residual xylitol dehydrogenase activity in the H. jecorina xdh1 deletion mutant.
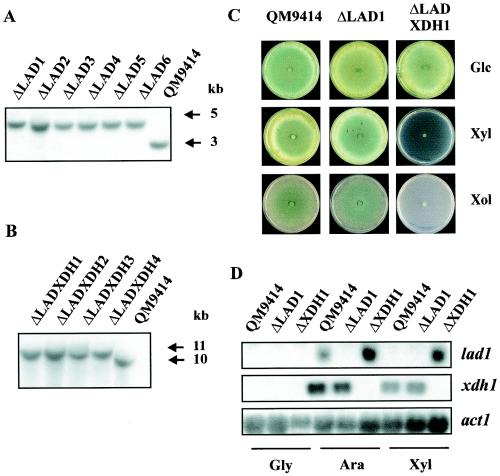
The results described above demonstrate that at least one more enzyme with xylitol dehydrogenase activity is present in H. jecorina and can partially compensate for the loss of Xdh1 function. A potential candidate for such an enzyme is lad1-encoded l-arabinitol-4-dehydrogenase, because it also catalyzes the oxidation of xylitol to xylulose (18). To study this possibility, the lad1 gene and its flanking regions were cloned and used to construct an xdh1 lad1 double-deletion strain of H. jecorina. First, H. jecorina lad1 deletion mutants were constructed by replacing the lad1 coding region with the H. jecorina pyr4 gene (Fig. 7A). The corresponding mutant strains were selected for growth on minimal medium with d-glucose as a carbon source and were tested for the disruption of lad1. Ten mutants were identified by Southern analysis to have undergone a single integration event in the lad1 locus. It is noteworthy that, in contrast to previous reports, the percentage of integration at the homologous locus was found to be about 50% for lad1. Second, to construct an xdh1 lad1 double-deletion strain of H. jecorina, the xdh1 coding region was replaced in an lad1 deletion strain as described above for strain ΔXDH1. Four transformants which showed the expected integration into the xdh1 locus (Fig. 7B) and in which the lad1 locus remained disrupted were obtained. The double-deletion mutant and the lad1 mutant were consequently tested for growth on d-xylose and xylitol (Fig. 7C). The data clearly show that the double-deletion strain lost the ability to grow on either of these carbon sources. Cell extracts from mycelia of strain ΔLADXDH1 that had been pregrown on glycerol and then transferred to d-xylose for 10 h did not contain any xylitol dehydrogenase activity, providing evidence that lad1 is responsible for the residual xylitol dehydrogenase activity still present in the xdh1 deletion mutant. In contrast, the lad1 single-deletion strain grew equally well on all of these carbon sources, demonstrating that the lad1 gene product is not involved in d-xylose and xylitol degradation in strains of H. jecorina when the xdh1 gene product is still functional.
FIG. 7.
Characterization of H. jecorina lad1 and lad1 xdh1 deletion mutants. (A) Southern analysis of lad1 deletion strains. Genomic DNAs of strain QM9414 and lad1 deletion strains were digested with BamHI and probed with a 1-kb BamHI-MluI fragment of lad1. In the lad1 deletion strains, replacement of the lad1 coding region by the H. jecorina pyr4 marker leads to a 1.7-kb increase in the size of the hybridizing fragment. This change leads to an increase in the size of the 3-kb hybridizing fragment in strain QM9414 to an ∼4.7-kb fragment in the lad1 deletion strains. (B) Southern analysis of lad1 xdh1 deletion strains, performed as described in the legend to Fig. 6A. (C) Comparison of the growth behaviors of parent strain QM9414 and deletion strains on plates containing d-glucose (Glc), d-xylose (Xyl), and xylitol (Xol) as carbon sources. (D) Northern analysis of lad1 and xdh1 transcript levels in strain QM9414 and deletion strains. Samples were obtained 6 h after transfer from a glycerol culture (Gly) to l-arabinose (Ara) and d-xylose (Xyl) cultures.
The compensation of xdh1 loss of function by the lad1 gene product is theoretically in conflict with the data of Richard et al. (18) showing that lad1 is not expressed on d-xylose. However, their data were obtained with carbon catabolite-derepressed strain RUT C-30. Therefore, we examined lad1 expression in the QM9414 background and in strain ΔXDH1. Consistent with the findings of Richard et al. (18), we did not find lad1 expression in strain QM9414, but we did find it in the xdh1 deletion strain (Fig. 7D). Therefore, lad1 is transcribed under conditions in which xdh1 function is impaired.
DISCUSSION
In the present work, we have characterized a xylitol dehydrogenase from the filamentous fungus H. jecorina at the molecular and functional levels. According to its primary structure, it belongs to the family of Zn2+-containing long-chain alcohol dehydrogenases, which also includes the xylitol, d-sorbitol, and iditol dehydrogenases of yeasts (8, 11, 17, 19). However, it differs from these enzymes in that it contains two predicted binding sites for Zn2+: in addition to a site which has three ligands typical for the binding of an active Zn2+ site (C50, H75, and E161) and which is conserved in all members of this enzyme family (8, 11, 17, 19), there is a second site (C105, C108, C111, and C119) which is not present in most of the other xylitol dehydrogenases (17). Therefore, it was concluded that the ligands of the second Zn2+ atom of the long-chain human alcohol dehydrogenase (C97, C100, C103, and C111) are not conserved in the yeast xylitol dehydrogenase, and it was suggested that this structural zinc atom is missing in all xylitol dehydrogenases and that this feature is characteristic of d-sorbitol dehydrogenases in general. However, the d-sorbitol dehydrogenase of S. pombe contains a second zinc binding site which is similar to that in H. jecorina, and the putative N. crassa xylitol dehydrogenase (NCU00891.1) also contains such a site. The phylogenetic analysis of xylitol and d-sorbitol dehydrogenases in this study showed that both the yeast and the filamentous fungal xylitol dehydrogenases arose from an S. pombe ancestor, implying that the enzymes in yeasts lost the second zinc binding site during evolution. That S. pombe is an evolutionary ancestor of Saccharomyces and Candida has also been evidenced by 28S gene sequence analysis (12). In addition, and despite containing two zinc binding sites, the H. jecorina xylitol dehydrogenase has the same substrate specificity as the yeast xylitol dehydrogenases (5, 13, 17, 19), namely, oxidation of xylitol and d-sorbitol, reduction of d-xylose and d-fructose, and no activity on l-arabinitol or l-arabinose. Although no extensive kinetic analysis was attempted, we conclude by analogy that this enzyme is a typical l-iditol:NAD+-2-oxidoreductase (EC 1.1.1.14).
The expression of xdh1 is adaptive, as no xdh1 transcript could be detected in mycelia grown on d-glucose or glycerol, whereas it accumulated during growth on d-xylose, xylitol, l-arabinose, and l-arabinitol. The effect of the latter two components may be direct or indirect, since the catabolism of l-arabinitol forms xylitol. The lack of xdh1 expression on d-glucose and glycerol would be indicative of regulation by carbon catabolite repression, but since the xdh1 transcript also does not accumulate during growth on glycerol in the cre1-truncated mutant H. jecorina RUT C-30 (unpublished data), this is clearly not the case. Therefore, the presence of the xdh1 transcript during growth on d-xylose is most likely due to induction, which would be consistent with the mode of regulation of the other two genes of the xylose catabolic pathway in A. niger (those for xylose reductase and xylulose-5-phosphate kinase) (9, 25). In addition, the Aspergillus xylulose-5-phosphate kinase was shown to be induced by l-arabinose and l-arabinitol but not by xylitol. As for the transactivator protein mediating this response, the A. niger xylose reductase was shown to be under the control of XlnR, the A. niger transcriptional activator of xylanase and cellulase biosynthesis (24, 26), whereas its xylulokinase was still inducible by d-xylose in an XlnR-negative mutant. Whether an XlnR homologue would also be involved in the regulation of xdh1 by d-xylose in H. jecorina is unclear, however, as we did not detect any nucleotide sequences matching the consensus sequence for the binding of XlnR (GGCTAA) in the xdh1 5′ upstream region. In H. jecorina, the genes encoding the two xylanases are differently expressed, and only xyn1 and not xyn2 is regulated by the XlnR homologue Xyr1 (14, 28). It is therefore possible that this coordinated regulation of genes for xylan and d-xylose metabolism in A. niger does not exist in H. jecorina, a possibility which may reflect the different natural habitats of the two fungi.
An intriguing finding of this study was that a loss-of-function mutant of xdh1 was still capable of growing on xylitol and—although at lower growth rates—on d-xylose, implying the presence of at least one more enzyme oxidizing xylitol or a less effective new pathway for d-xylose catabolism. The results obtained with the xdh1 lad1 double-deletion mutant conclusively showed that the enzyme responsible for this residual activity is l-arabinitol-4-dehydrogenase, Lad1. The lack of both Xdh1 and Lad1 leads to a complete loss of the ability to grow on d-xylose and xylitol. Lad1 and Xdh1 belong to the same family of zinc-containing alcohol dehydrogenases, and a phylogenetic analysis of Xdh1 and Lad1 from various sources showed that the clade containing Lad1 is basal to that of Xdh1; these data imply that the more specific Xdh1 protein may have evolved from the rather broadly specific Lad1 protein. Such an assumption is also supported by the finding that H. jecorina xdh1 was not able to enable a loss-of-function mutant of lad1 to grow on l-arabinose, consistent with the inability of Xdh1 to oxidize l-arabinitol. Compensation of loss-of-function mutants in one pentose catabolic pathway by enzymes from another pathway may also explain the results obtained with A. niger, i.e., that a xylose reductase mutant was still able to grow at a reduced rate on d-xylose (9). The pathways for d-xylose and l-arabinose catabolism are both initiated by an aldose reductase, but at present it is not known whether the same enzyme, two isoenzymes with the same substrate specificity, or two enzymes with different substrate specificities catalyze the steps in the two pathways.
Despite of the lack of l-arabinitol-oxidizing activity of Xdh1, the xdh1 mutant showed a significantly decreased growth rate on l-arabinitol, an intriguing result in view of the lack of effect on growth on xylitol in this mutant. Although we cannot rule out the possibility that the xdh1 gene product also has a regulatory function, we interpret the findings in terms of an imbalance in substrates, products, and/or coenzymes such that flux like that in the wild type cannot be maintained. The fact that these findings are observed only when the first (NADPH-specific) step (l-arabinose reductase) is omitted suggests that the NADPH/NADP ratio may be the critical variable: filamentous fungi possess a cytosolic pyridine nucleotide transhydrogenase (e.g., N. crassa genome database entry NCU01140.1; http://www-genome.wi.mit.edu/annotation/fungi/neurospora) which is responsible for maintaining a balance between NADH/NAD and NADPH/NADP ratios. The catabolism of l-arabinose to l-xylulose-5-phosphate requires two NADPH and two NAD+ molecules, and it is likely that the transhydrogenase is at least partially involved in the regeneration of NAD+ for the l-arabinitol-4-dehydrogenase and xylitol dehydrogenase reactions through reduction of the NADP+ formed in the aldose reductase reactions. Thus, under conditions where only l-arabinitol-4-dehydrogenase accounts for pentitol reductions, the regeneration of NAD+ on behalf of only one NADPH molecule generated by the d-xylulose reductase reaction located downstream may not produce sufficient activity to catalyze l-arabinitol oxidation with the same velocity as in the wild type. This interpretation is also supported by the generally lower growth rate of the wild type on both l-arabinitol and xylitol. However, the proposed role of the pyridine nucleotide transhydrogenase needs to be verified by reverse genetics.
Acknowledgments
This study was supported by a grant from the Austrian Science Foundation (P15131-MOB) to C.P.K., which is gratefully acknowledged.
Lukas Hartl and Manuela Pail contributed equally to this work.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2003. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 3.Chakravorty, M., and B. C. Horecker. 1966. Polyol dehydrogenases of Candida utilis. I. DPN-linked dehydrogenase. Methods Enzymol. 9:163-165. [Google Scholar]
- 4.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditzelmüller, G., C. P. Kubicek, W. Wöhrer, and M. Röhr. 1984. Xylitol dehydrogenase from Pachysolen tannophilus. FEMS Microbiol. Lett. 25:195-198. [Google Scholar]
- 6.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaf. 1990. Cloning of the Trichoderma reesei pyrG-gene and its use as a homologous marker for a high-frequency transformation system. Curr. Genet. 18:447-451. [DOI] [PubMed] [Google Scholar]
- 7.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaff. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18:71-76. [DOI] [PubMed] [Google Scholar]
- 8.Habenicht, A., H. Motejadded, M. Kiess, A. Wegerer, and R. Mattes. 1999. Xylose utilisation: cloning and characterisation of the xylitol dehydrogenase from Galactocandida mastotermitis. Biol. Chem. 380:1405-1411. [DOI] [PubMed] [Google Scholar]
- 9.Hasper, A. A., J. Visser, and L. H. de Graaff. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 36:193-200. [DOI] [PubMed] [Google Scholar]
- 10.Jeffries, T. W., and N. Q. Shi. 1999. Genetic engineering for improved xylose fermentation by yeasts. Adv. Biochem. Eng. Biotechnol. 65:117-161. [DOI] [PubMed] [Google Scholar]
- 11.Kotter, P., R. Amore, C. P. Hollenberg, and M. Ciriacy. 1990. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr. Genet. 18:493-500. [DOI] [PubMed] [Google Scholar]
- 12.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 13.Lunzer, R., Y. Mamnun, D. Haltrich, K. D. Kulbe, and B. Nidetzky. 1998. Structural and functional properties of a yeast xylitol dehydrogenase, a Zn2+-containing metalloenzyme similar to medium-chain sorbitol dehydrogenases. Biochem. J. 336:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mach, R. L., and S. Zeilinger. 2003. Regulation of gene expression in industrial fungi: Trichoderma. Appl. Microbiol. Biotechnol. 60:515-522. [DOI] [PubMed] [Google Scholar]
- 15.Mach, R. L., M. Schindler, and C. P. Kubicek. 1994. Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr. Genet. 25:567-570. [DOI] [PubMed] [Google Scholar]
- 16.Mandels, M. M., and R. E. Andreotti. 1978. The cellulose to cellulase fermentation. Proc. Biochem. 13:6-13. [Google Scholar]
- 17.Persson, B., J. Hallborn, M. Walfridsson, B. Hahn-Hagerdal, S. Keranen, M. Penttila, and H. Jornvall. 1993. Dual relationships of xylitol and alcohol dehydrogenases in families of two protein types. FEBS Lett. 324:9-14. [DOI] [PubMed] [Google Scholar]
- 18.Richard, P., J. Londesborough, M. Putkonen, N. Kalkkinen, and M. Penttila. 2001. Cloning and expression of a fungal l-arabinitol 4-dehydrogenase gene. J. Biol. Chem. 276:40631-40637. [DOI] [PubMed] [Google Scholar]
- 19.Richard, P., M. H. Toivari, and M. Penttila. 1999. Evidence that the gene YLR070c of Saccharomyces cerevisiae encodes a xylitol dehydrogenase. FEBS Lett. 457:135-138. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Seiboth, B., G. Hofmann, and C. P. Kubicek. 2002. Lactose metabolism and cellulase production in Hypocrea jecorina: the gal7 gene, encoding galactose-1-phosphate uridylyltransferase, is essential for growth on galactose but not for cellulase induction. Mol. Genet. Genomics 267:124-132. [DOI] [PubMed] [Google Scholar]
- 22.Slayman, C. W., and E. L. Tatum. 1964. Potassium transport in Neurospora. I. Intracellular sodium and potassium concentrations, and cation requirements for growth. Biochim. Biophys. Acta 88:578-592. [PubMed] [Google Scholar]
- 23.Timell, T. E. 1965. Wood hemicelluloses. Part II. Adv. Carbohydr. Chem. 20:409-483. [PubMed] [Google Scholar]
- 24.vanKuyk, P. A., M. J. de Groot, G. J. Ruijter, R. P. de Vries, and J. Visser. 2001. The Aspergillus niger d-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, and is essential for growth on d-xylose and l-arabinose. Eur. J. Biochem. 268:5414-5423. [DOI] [PubMed] [Google Scholar]
- 25.van Peij, N. N., M. M. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Peij, N. N., J. Visser, and L. H. de Graaff. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131-142. [DOI] [PubMed] [Google Scholar]
- 27.Wang, T., M. Penttilä, P. Gao, C. Wang, and L. Zhong. 1998. Isolation and identification of xylitol dehydrogenase gene from Trichoderma reesei. Chin. J. Biotechnol. 14:179-185. [PubMed] [Google Scholar]
- 28.Würleitner, E., L. Pera, C. Wacenovsky, A. Cziferszky, S. Zeilinger, C. P. Kubicek, and R. L. Mach. 2003. Transcriptional regulation of xyn2 in Hypocrea jecorina. Eukaryot. Cell 2:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]