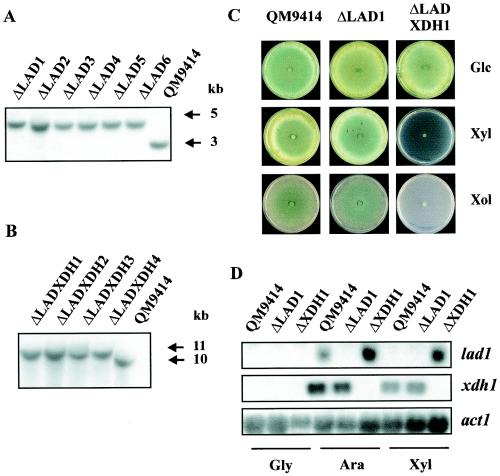
FIG. 7.
Characterization of H. jecorina lad1 and lad1 xdh1 deletion mutants. (A) Southern analysis of lad1 deletion strains. Genomic DNAs of strain QM9414 and lad1 deletion strains were digested with BamHI and probed with a 1-kb BamHI-MluI fragment of lad1. In the lad1 deletion strains, replacement of the lad1 coding region by the H. jecorina pyr4 marker leads to a 1.7-kb increase in the size of the hybridizing fragment. This change leads to an increase in the size of the 3-kb hybridizing fragment in strain QM9414 to an ∼4.7-kb fragment in the lad1 deletion strains. (B) Southern analysis of lad1 xdh1 deletion strains, performed as described in the legend to Fig. 6A. (C) Comparison of the growth behaviors of parent strain QM9414 and deletion strains on plates containing d-glucose (Glc), d-xylose (Xyl), and xylitol (Xol) as carbon sources. (D) Northern analysis of lad1 and xdh1 transcript levels in strain QM9414 and deletion strains. Samples were obtained 6 h after transfer from a glycerol culture (Gly) to l-arabinose (Ara) and d-xylose (Xyl) cultures.