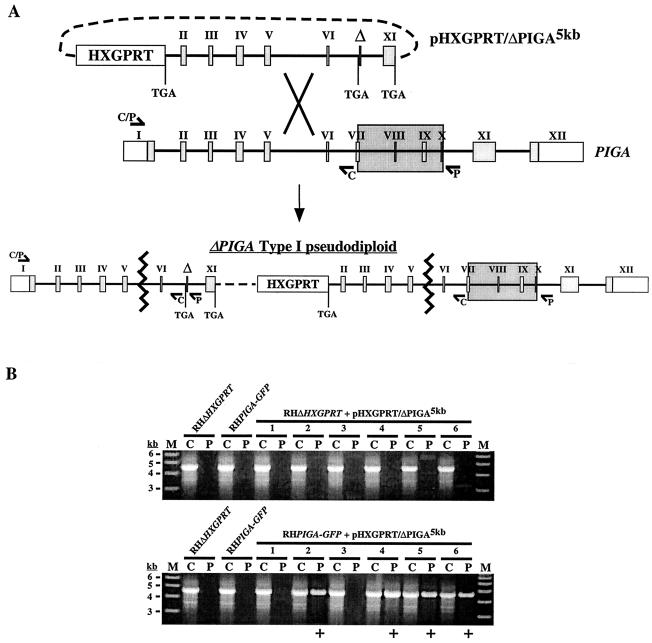
FIG. 3.
Targeted disruption of PIGA. (A) Schematic of the gene disruption strategy. The pHXGPRT/ΔPIGA5kb knockout vector was constructed such that single-crossover homologous recombination into the PIGA locus upstream of the deletion would generate a type I pseudodiploid with two nonfunctional PIGA alleles. To generate ΔPIGA5kb, the 5′ end of PIGA was truncated within exon 1 (removing the 5′ untranslated region and coding sequence corresponding to residues 1 to 47) and the 3′ end was truncated into exon 11 (removing the coding sequence for the putative transmembrane and ER-lumenal domains [residues 519 to 616]). An internal sequence corresponding to a highly conserved region of PIGA (amino acids 303 to 368) was also deleted, and stop codons were engineered into all cloning junctions within the ΔPIGA5kb allele. Type I integration resulted in the harboring by the pairs upstream allele of the ∼1.8-kb deletion (marked by Δ; large shaded box indicates the corresponding sequence in the wild-type allele), which could be detected by PCR (using primer set P) as a product of ∼4.6 kb. The P primer set was found to be incapable of amplifying the predicted product (∼6.5 kb) from wild-type genomic DNA. (B) Six independent stable transgenic populations (lane pairs 1 to 6) derived from either parental parasites (RHΔHXGPRT) or parasites harboring a stable second copy of PIGA (RHPIGA-GFP) were screened for integration of the plasmid into PIGA (see text and elsewhere in this legend). ΔPIGA type I pseudodiploids were not detected in any of the six populations derived from parental parasites, while four out of six populations from RHPIGA-GFP parasites yielded a positive PCR product result (+). Lanes C, control primers; lanes P, pseudodiploid-specific primers; lanes M, molecular mass markers.